- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Engineered DNA-binding Molecule-mediated Chromatin Immunoprecipitation (in vitro enChIP) Using CRISPR Ribonucleoproteins in Combination with Next-generation Sequencing (in vitro enChIP-Seq) for the Identification of Chromosomal Interactions
Published: Vol 7, Iss 22, Nov 20, 2017 DOI: 10.21769/BioProtoc.2612 Views: 10429
Reviewed by: Jihyun KimXiao LiKate Hannan

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Construction of DNA/RNA Triplex Helices Based on GAA/TTC Trinucleotide Repeats
Jiahui Zhang [...] Celeste Sagui
Sep 20, 2021 3567 Views

Quantification of Chromosomal Aberrations in Mammalian Cells
Inés Paniagua and Jacqueline J. L. Jacobs
Aug 20, 2023 2838 Views

Electrophoretic Mobility Assay to Separate Supercoiled, Catenated, and Knotted DNA Molecules
Jorge Cebrián [...] María José Fernández-Nestosa
May 5, 2024 2152 Views
Abstract
We have developed locus-specific chromatin immunoprecipitation (locus-specific ChIP) technologies consisting of insertional ChIP (iChIP) and engineered DNA-binding molecule-mediated ChIP (enChIP). Locus-specific ChIP is a method to isolate a genomic region of interest from cells while it also identifies what binds to this region using mass spectrometry (for protein) or next generation sequencing (for RNA or DNA) as described in Fujita et al. (2016a). Recently, we identified genomic regions that physically interact with a locus using an updated form of enChIP, in vitro enChIP, in combination with NGS (in vitro enChIP-Seq) (Fujita et al., 2017a). Here, we describe a protocol on in vitro enChIP to isolate a target locus for identification of genomic regions that physically interact with the locus.
Keywords: Chromatin immunoprecipitationBackground
Elucidation of molecular mechanisms underlining genome functions requires the identification of molecules that interact with the genomic region of interest in vivo. To this end, we have developed locus-specific chromatin immunoprecipitation (locus-specific ChIP) technologies consisting of insertional ChIP (iChIP) and engineered DNA-binding molecule-mediated ChIP (enChIP) (Fujita et al., 2016a). Locus-specific ChIP is a method to biochemically isolate a genomic region of interest from cells. Molecules interacting with that isolated genomic region are identified by biochemical analyses, such as mass spectrometry (MS) and next generation sequencing (NGS). In iChIP, an exogenous DNA-binding protein and its recognition DNA sequence are used for ‘in cell’ locus-tagging. In enChIP, engineered DNA-binding molecules, such as transcription activator-like (TAL) proteins and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), are used for ‘in cell’ locus-tagging. The tagged loci are isolated by affinity-purification. We recently developed in vitro enChIP, in which the locus-tagging was performed in vitro (in a test tube) using recombinant and/or synthetic engineered DNA-binding molecules (Figure 1) (Fujita and Fujii, 2014a and 2016b). Here, we describe a protocol of in vitro enChIP using CRISPR ribonucleoproteins (RNPs) followed by NGS (in vitro enChIP-Seq) for the identification of genomic regions that physically interact with the locus of interest (Fujita et al., 2017a).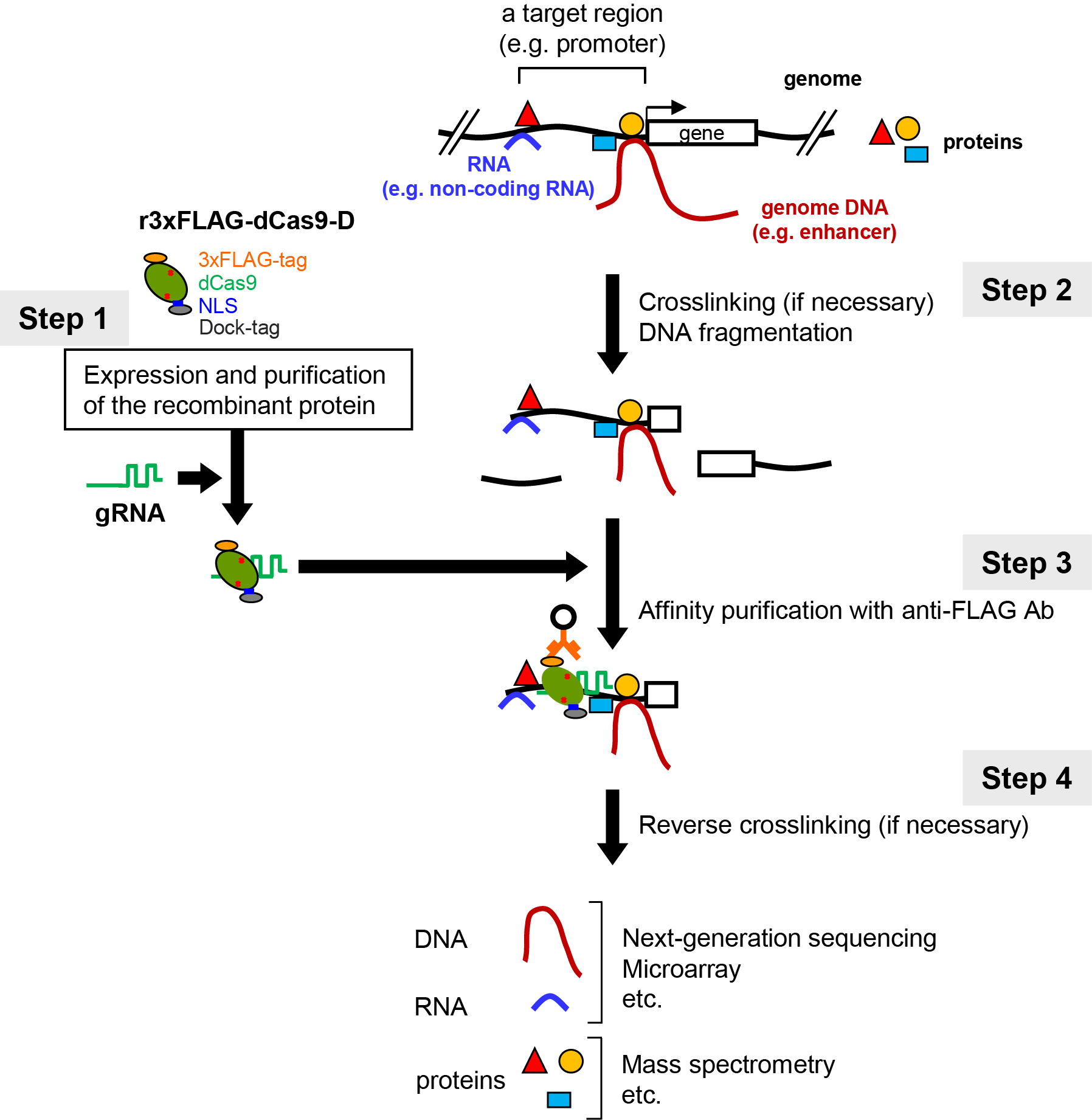
Figure 1. Scheme of in vitro enChIP using CRISPR RNPs. Figure 1 reproduced under the Creative Commons Attribution License from: Fujita et al. Efficient sequence-specific isolation of DNA fragments and chromatin by in vitro enChIP technology using recombinant CRISPR ribonucleoproteins. Genes Cells. 2016; 21: 370-377.
Scheme of in vitro enChIP using CRISPR RNPs (Figure 1)
1. A nuclease-dead form of Cas9 (dCas9) fused to an epitope-tag(s) (e.g., 3xFLAG-dCas9) is prepared as a recombinant protein. Guide RNA (gRNA) that recognizes the DNA sequence of the genomic region of interest is generated chemically. As to gRNA, single gRNA (sgRNA) or a complex of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) can be used.
2. Cells to be analyzed are crosslinked, if necessary, and lysed, and DNA is fragmented.
3. The 3xFLAG-dCas9/gRNA complex is incubated with the fragmented chromatin DNA in a test tube. The genomic DNA bound to the 3xFLAG-dCas9/gRNA complex is affinity-purified using antibody against the epitope-tag(s) or dCas9 itself. Alternatively, the complex can be purified using tags fused to gRNA (e.g., biotin). The isolated complexes retain molecules that interact with the target locus.
4. Reverse crosslinking, if necessary, and subsequent purification of DNA, RNA, or proteins allows identification and characterization of these molecules. For example, NGS or MS can be combined to identify genomic regions or proteins that physically bind to the target locus.
We previously described a detailed protocol of ‘in cell’ enChIP for the identification of proteins that interact with a locus of interest in Bio-protocol (Fujita and Fujii, 2014b). Procedures for the preparation of sonicated chromatin in in vitro enChIP are the same as those for ‘in cell’ enChIP. In addition, other procedures (e.g., preparation of Dynabeads) are similar between them. Therefore, we recommend to refer to the ‘in cell’ enChIP protocol (Fujita and Fujii, 2014b) in parallel.
Materials and Reagents
- 1.5 ml centrifuge tube (SARSTEDT, catalog number: 72.690.001 )
- Pipettes tips (DNase/RNase-free)
- DT40 cell (RIKEN BioResource Center) (an example)
- 10 µM crRNA (FASMAC, diluted in DNase/RNase-free water) (see step B1)
- 10 µM tracrRNA (FASMAC, diluted in DNase/RNase-free water) (see step B1)
- 3xFLAG-dCas9-D (Sysmex, ProCube)
- 37% formaldehyde (NACALAI TESQUE, catalog number: 16223-55 )
- Anti-FLAG M2 antibody (Sigma-Aldrich, catalog number: F1804 )
- Normal mouse IgG (Santa Cruz Biotechnology, catalog number: sc-2025 )
- Dynabeads-Protein G (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10004D )
- RNasin plus RNase inhibitor (Promega, catalog number: N261A )
- 10 mg/ml RNase A (Sigma-Aldrich, catalog number: R6513 )
- 10% SDS solution (NACALAI TESQUE, catalog number: 30562-04 )
- 20 mg/ml Proteinase K (Roche Diagnostics, catalog number: 3115828001 )
- ChIP DNA Clean and Concentrator (ZYMO RESEARCH, catalog number: D5205 )
- TruSeq ChIP Sample Prep Kit (Illumina, catalog number: IP-202-1012 )
- Potassium chloride (KCl) (NACALAI TESQUE, catalog number: 28538-62 )
- Dithiothreitol (DTT) (NACALAI TESQUE, catalog number: 14128-91 )
- 1 M HEPES (pH 7.1-7.5) (NACALAI TESQUE, catalog number: 17557-94 )
- 0.5 M ethylenediaminetetraacetate acid (EDTA) (pH 8.0) (NACALAI TESQUE, catalog number: 06894-85 )
- 1 M magnesium chloride hexahydrate (MgCl2·6H2O) (NACALAI TESQUE, catalog number: 20942-34 )
- cOmplete, Mini, EDTA-free Protease Inhibitor (Roche Diagnostics, catalog number: 4693159001 )
- UltraPure DNase/RNase-Free Distilled Water (DDW) (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10977015 )
- 10x phosphate buffered saline (PBS) (pH 7.4) (NACALAI TESQUE, catalog number: 27575-31 )
- 1x PBS (10x dilution of 10x PBS with distilled water)
- Tween-20 (Sigma-Aldrich, catalog number: P5927 )
Note: This product has been discontinued. - BSA fraction V (7.5%) (Thermo Fisher Scientific, catalog number: 15260037 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9625 )
- 1 M Tris (pH 8.0) (AppliChem, catalog number: A4577 )
- 1 M Tris (pH 7.5) (AppliChem, catalog number: A4263 )
- Polyethylene Glycol Mono-p-isooctylphenyl Ether (Triton X-100) (NACALAI TESQUE, catalog number: 12967-45 )
- IGEPAL CA-630 (Sigma-Aldrich, catalog number: I8896 )
- 3xFLAG peptide (Sigma-Aldrich, catalog number: F4799 )
- 1 M KCl (see Recipes)
- 0.1 M DTT (see Recipes)
- In vitro CRISPR buffer (+ 0.5 mM DTT) (see Recipes)
- PBS-T (see Recipes)
- PBS-T-BSA (see Recipes)
- 5 M NaCl (see Recipes)
- Modified low salt buffer (+ 0.04% SDS) (see Recipes)
- TBS (see Recipes)
- TBS-IGEPAL CA-630 (see Recipes)
- Elution buffer (see Recipes)
Equipment
- Pipettes
- Ultrasonic Disruptor (TOMY SEIKO, model: UD-201 )
- Block heater (Eppendorf, catalog number: 5355 000.046 )
- Magnetic stand (Magical Trapper) (TOYOBO, catalog number: MGS-101 )
- Centrifuge (Eppendorf, catalog number: 5427 R )
- Vortex mixer (Fisher Scientific, catalog number: 128101 )
- Rotator (AS ONE, catalog number: TR-118 )
- HiSeq 2500 system (Illumina, model: HiSeq 2500 System )
Software
- Integrative Genomics Viewer, http://software.broadinstitute.org/software/igv/IGV
Procedure
- Preparation of sonicated chromatin
- Formaldehyde crosslinking of cells (e.g., 2 x 107 DT40 cells, 1% formaldehyde in culture medium at 37 °C for 5 min) and preparation of chromatin are performed as described previously (steps B and C in Fujita and Fujii, 2014b).
- Sonicate the prepared chromatin (e.g., Output: 3, Duty: 100%, Time: Free, 10 sec x 15 cycles using Ultrasonic Disruptor UD-201 (TOMY)) as described previously (step D in Fujita and Fujii, 2014b) except for substituting 800 µl of in vitro CRISPR buffer (+ 0.5 mM DTT) (see Recipes) instead of MLB3.
- Evaluate fragmentation of chromatin (the average length of fragments is about 2 kbp) as described previously (step E in Fujita and Fujii, 2014b).
- Formaldehyde crosslinking of cells (e.g., 2 x 107 DT40 cells, 1% formaldehyde in culture medium at 37 °C for 5 min) and preparation of chromatin are performed as described previously (steps B and C in Fujita and Fujii, 2014b).
- Preparation of gRNA
- Mix 2 µl of 10 µM crRNA and 2 µl of 10 µM tracrRNA. In our previous study (Fujita et al., 2017a), a crRNA (cggcaggcucgggugcgccuguuuuagagcuaugcuguuuug) and a tracrRNA (aacagcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucggugcuuuuuuu) were used for targeting the chicken Pax5 promoter region; the underlined sequence is corresponding to the target site.
- Incubate at 100 °C for 2 min in a block heater. Incubation for 2 min does not make considerable evaporation. After heating, put the RNAs out of the heater to naturally cool down to room temperature. The RNAs are ready for step D6.
- Mix 2 µl of 10 µM crRNA and 2 µl of 10 µM tracrRNA. In our previous study (Fujita et al., 2017a), a crRNA (cggcaggcucgggugcgccuguuuuagagcuaugcuguuuug) and a tracrRNA (aacagcauagcaaguuaaaauaaggcuaguccguuaucaacuugaaaaaguggcaccgagucggugcuuuuuuu) were used for targeting the chicken Pax5 promoter region; the underlined sequence is corresponding to the target site.
- Preparation of Dynabeads conjugated with antibody (anti-FLAG antibody or normal mouse IgG)
- Prepare two 1.5 ml tubes; one is for anti-FLAG antibody and the other is for normal mouse IgG. Add 20 µl slurry of Dynabeads-Protein G in each tube (20 µl x 2 tubes). In this regard, we usually use 20 µl slurry of the beads for 2 µg of an antibody (see step C7). In this protocol, we describe preparation of one sample for NGS analysis. In this regard, normal mouse IgG is used for pre-clear in step E1 and anti-FLAG antibody is used in Procedure D. If you handle another sample (e.g., affinity purification without gRNA as a negative control), you may prepare chromatin and Dynabeads for it separately.
- Place the tubes on the magnetic stand on ice for 3 min. Discard the supernatant by pipetting. Detailed instructions on the use of the magnetic stand are illustrated in Figure 2 in Fujita and Fujii, 2014b.
- Resuspend the pellet in 0.5 ml of PBS-T (see Recipes). Place the tubes on the magnetic stand on ice for 3 min. Discard the supernatant by pipetting.
- Repeat step F3 (total of twice).
- Resuspend the pellet in 0.5 ml of PBS-T-BSA (see Recipes).
- Add 2 µg of antibody, to the beads either anti-FLAG or normal mouse IgG. Rotate at 4 °C overnight. As to Dynabeads conjugated with anti-FLAG antibody, proceed to Procedure D. As to Dynabeads conjugated with normal mouse IgG, proceed to the next step (C7).
- Centrifuge briefly (400 x g for 2-3 sec). Place the tubes on the magnetic stand on ice for 3 min. Discard the supernatant by pipetting.
- Resuspend the pellet in 0.5 ml of PBS-T. Invert several times and centrifuge briefly (400 x g for 2-3 sec). Place the tubes on the magnetic stand on ice for 3 min. Discard the supernatant by pipetting.
- Repeat step C8, twice (total of three times). The Dynabeads conjugated with normal mouse IgG are ready for step E1.
- Prepare two 1.5 ml tubes; one is for anti-FLAG antibody and the other is for normal mouse IgG. Add 20 µl slurry of Dynabeads-Protein G in each tube (20 µl x 2 tubes). In this regard, we usually use 20 µl slurry of the beads for 2 µg of an antibody (see step C7). In this protocol, we describe preparation of one sample for NGS analysis. In this regard, normal mouse IgG is used for pre-clear in step E1 and anti-FLAG antibody is used in Procedure D. If you handle another sample (e.g., affinity purification without gRNA as a negative control), you may prepare chromatin and Dynabeads for it separately.
- Preparation of Dynabeads conjugated with recombinant CRISPR RNPs
- Add 2 µg of recombinant 3xFLAG-dCas9-D to the tube, in which Dynabeads are conjugated with anti-FLAG antibody in 0.5 ml of PBS-T-BSA (step C6). Rotate at 4 °C for 2-3 h.
- Centrifuge briefly (400 x g for 2-3 sec). Place the tubes on the magnetic stand on ice for 3 min. Discard the supernatant by pipetting.
- Resuspend the pellet in 0.5 ml of PBS-T. Invert several times and centrifuge briefly (400 x g for 2-3 sec). Place the tubes on the magnetic stand on ice for 3 min. Discard the supernatant by pipetting.
- Repeat step D3 (total of twice).
- Resuspend the pellet in 0.5 ml of in vitro CRISPR buffer (+ 0.5 mM DTT). Invert several times and centrifuge briefly (400 x g for 2-3 sec). Place the tubes on the magnetic stand on ice for 3 min. Discard the supernatant by pipetting.
- Add 100 µl of in vitro CRISPR buffer (+ 0.5 mM DTT) and 4 µl of the gRNA prepared in Procedure B.
- Incubate at 37 °C for 10 min. The Dynabeads conjugated with recombinant CRISPR RNPs are ready for step E3.
- Add 2 µg of recombinant 3xFLAG-dCas9-D to the tube, in which Dynabeads are conjugated with anti-FLAG antibody in 0.5 ml of PBS-T-BSA (step C6). Rotate at 4 °C for 2-3 h.
- Affinity purification (chromatin immunoprecipitation)
- Take 800 µl of the fragmented chromatin (Procedure A), which corresponds to chromatin extracted from 2 x 107 cells, and add to the tube containing the Dynabeads conjugated with normal mouse IgG (step C9). Rotate at 4 °C for 1 h.
- Place the tubes on the magnetic stand on ice for 3 min.
- Transfer the cleared supernatant into the tube, in which the Dynabeads conjugated with recombinant CRISPR RNPs were prepared (step D7). Add RNase inhibitor (40 U/ml, final concentration).
- Incubate at 37 °C for 20 min at 650 rpm in a block heater.
- Place the tubes on the magnetic stand on ice for 3 min. Discard the supernatant by pipetting.
- Wash the beads with 1 ml of modified low salt buffer (+ 0.04% SDS) (see Recipes). Rotate at 4 °C for 10 min. Place the tubes on the magnetic stand on ice for 3 min. Discard the supernatant by pipetting.
- Repeat step E6, three times (total of 4 times).
- Add 1 ml of TBS-IGEPAL CA-630 (see Recipes). Rotate at 4 °C for 10 min. Place the tubes on the magnetic stand on ice for 3 min. Discard the supernatant by pipetting.
- Elution 1: Resuspend the beads in 60 µl of elution buffer (see Recipes). Incubate at 37 °C for 20 min at 850 rpm in a block heater. Place the tubes on the magnetic stand on ice for 3 min. Transfer the supernatant into a 1.5 ml tube.
- Elution 2: Resuspend the beads in 40 µl of elution buffer. Incubate at 37 °C for 5 min at 850 rpm in a block heater. Place the tubes on the magnetic stand on ice for 3 min. Transfer the supernatant into the 1.5 ml tube, in which 60 µl of the eluate was collected (total 100 µl).
- Add 4 μl of 5 M NaCl (see Recipes). Incubate at 65 °C overnight.
- Add 2 µl of 10 mg/ml RNase A. Incubate at 37 °C for 1 h.
- Add 5 µl of 10% SDS and 5 µl of 20 mg/ml Proteinase K. Incubate at 45 °C for 2 h.
- Purify DNA with ChIP DNA Clean and Concentrator (Zymo Resarch) according to the manufacturer’s protocol (e.g., 0.6 ml of ChIP DNA binding buffer and 60 µl of elution buffer in the kit can be used).
- Take 800 µl of the fragmented chromatin (Procedure A), which corresponds to chromatin extracted from 2 x 107 cells, and add to the tube containing the Dynabeads conjugated with normal mouse IgG (step C9). Rotate at 4 °C for 1 h.
- Preparation of NGS libraries and NGS analysis
- Prepare NGS libraries from the DNA samples (step E14) using TruSeq ChIP Sample Prep Kit (Illumina) according to the manufacturer’s protocol.
- Subject the DNA libraries to DNA sequencing using the HiSeq 2500 system (36 bp single end, 20-50 million reads).
- You can visualize the NGS data in an appropriate viewer (e.g., Integrative Genomics Viewer, http://software.broadinstitute.org/software/igv/IGV) (Figures 2 and 3).
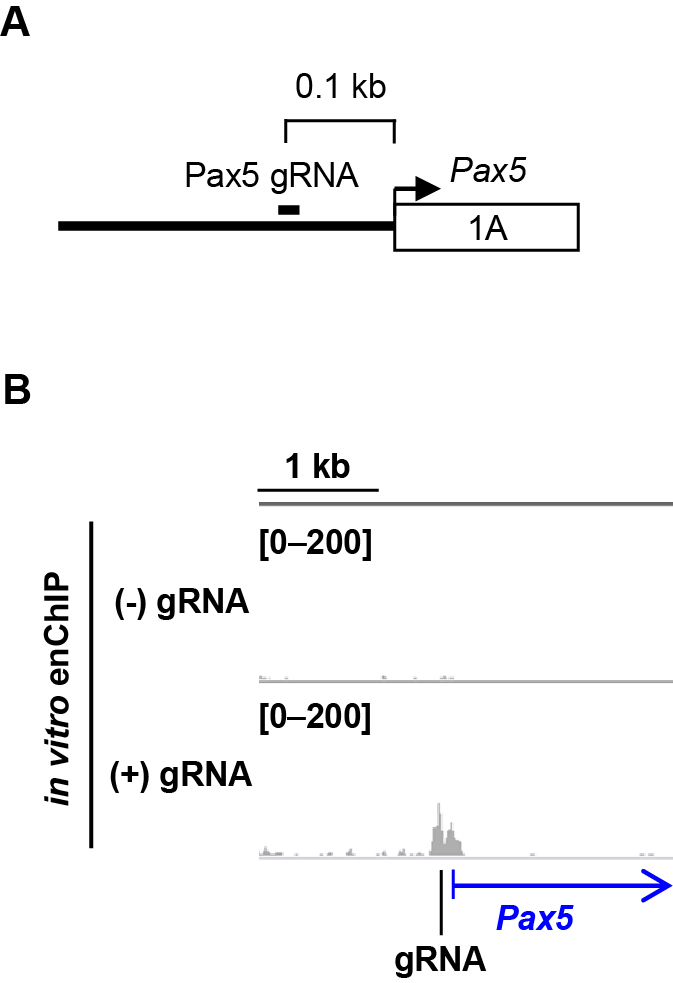
Figure 2. Isolation of a target genomic region by in vitro enChIP. A. Target position of a gRNA; B. NGS peak images around the target region. NGS data were visualized in IGV. Figure 2 reproduced under the Creative Commons Attribution License from: Fujita et al. Locus-specific ChIP combined with NGS analysis reveals genomic regulatory regions that physically interact with the Pax5 promoter in a chicken B cell line. DNA Res. 2017, 24: 537-548.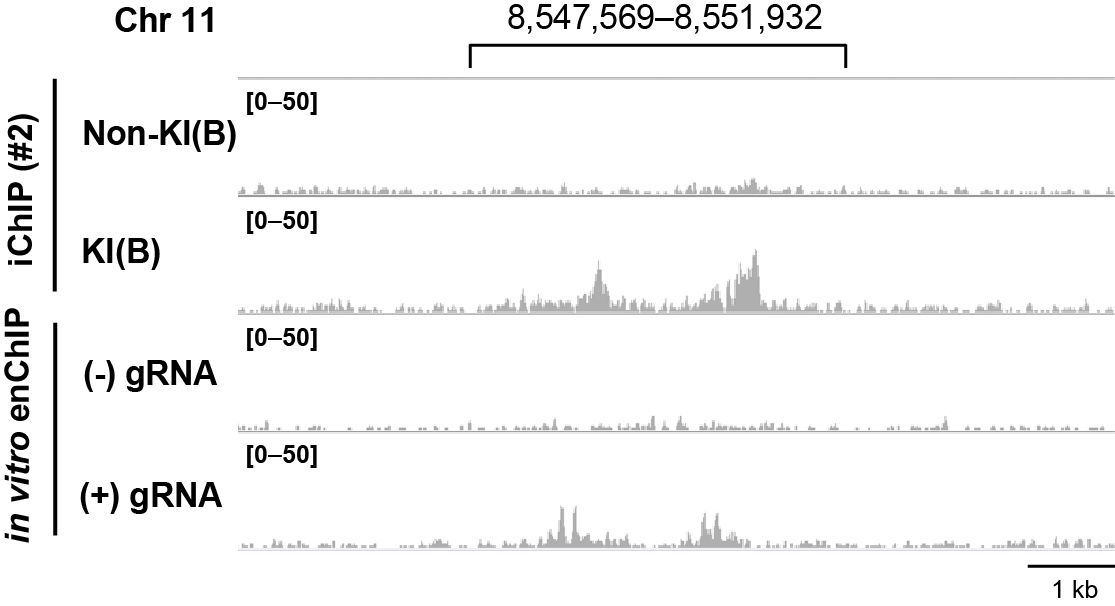
Figure 3. Identification of genomic regions interacting with the Pax5 promoter region. Genomic regions that physically interact with the Pax5 promoter region. NGS data were visualized in IGV. Figure 3 reproduced under the Creative Commons Attribution License from: Fujita et al. Locus-specific ChIP combined with NGS analysis reveals genomic regulatory regions that physically interact with the Pax5 promoter in a chicken B cell line. DNA Res. 2017, 24: 537-548.
- Prepare NGS libraries from the DNA samples (step E14) using TruSeq ChIP Sample Prep Kit (Illumina) according to the manufacturer’s protocol.
Data analysis
- For visualization in IGV, the NGS data (BAM file) is uploaded to IGV. A reference genome information (e.g., galGal4) is also loaded to map the NGS data.
- Enlarge the position of the target genomic region in the browser to confirm isolation of a target genomic region (Figure 2B). Also check the NGS data with a negative control NGS data (e.g., in vitro enChIP-Seq without gRNA) to judge if the signal at the target genomic region is not noise.
- Viewing on a genome-wide scale, you can manually identify genomic regions that interact with the target genomic region (Figure 3). In this regard, the peak regions detected in the NGS data but not in a negative control NGS data can be judged as interacting genomic regions. Narrow peaks of each in vitro enChIP-Seq dataset can be automatically detected using MACS2 or other software (Fujita et al., 2017b).
Notes
- It is also possible to combine conventional ‘in cell’ enChIP using CRISPR with NGS (enChIP-Seq) (Fujita et al., 2017b). In this case, the CRISPR components (e.g., 3xFLAG-dCas9 and sgRNA) are expressed in cells and enChIP is performed to isolate a target locus as described previously (Fujita and Fujii, 2014b and 2015). After isolation of the locus, DNA is purified and analyzed as shown in steps E12-F3.
- It is possible to confirm isolation of the target genomic region by qPCR before NGS analysis. To this end, the DNA sample (step E14) is subjected to qPCR with a primer set specific to the target genomic region. Examples are shown previously (Fujita et al., 2016b and 2017a).
- Off-target binding sites may be listed as candidates of interacting genomic regions. To eliminate such potential off-target sites, it would be necessary to check whether DNA sequences of the candidate genomic regions do not contain sequences similar to the target sequences of gRNA.
- To increase the reliability of extraction of interacting genomic regions, it would be useful to perform in vitro enChIP with two or more gRNAs to the same genomic region. In this regard, genomic regions commonly detected by in vitro enChIP using different gRNAs (except for the target genomic region) can be judged as bona fide interacting genomic regions. Details are described previously (Fujita et al., 2017b)
Recipes
- 1 M KCl solution (50 ml)
3.73 g KCl (MW: 74.55)
DDW to 50 ml - 0.1 M DTT solution (10 ml) (store at -20 °C)
0.15 g DTT (MW: 154.25)
DDW to 10 ml - in vitro CRISPR buffer (+ 0.5 mM DTT)
20 mM HEPES, pH 7.1-7.5
150 mM KCl
0.1 mM EDTA
10 mM MgCl2
0.5 mM DTT
1x protease inhibitors
For preparing 50 ml:
1 ml 1 M HEPES (pH 7.1-7.5)
7.5 ml 1 M KCl
10 µl 0.5 M EDTA
500 µl 1 M MgCl2
250 µl 0.1 M DTT (add just before use)
5 tablets cOmplete, Mini, EDTA-free (add just before use)
41 ml DDW - PBS-T (10 ml)
PBS (pH 7.4), 0.01% Tween-20
10 ml PBS
10 µl 10% Tween-20 - PBS-T-BSA
PBS (pH 7.4)
0.01% Tween-20
0.1% BSA
For preparing 10 ml:
10 ml PBS
10 µl 10% Tween-20
133 µl 7.5% BSA fraction V - 5 M NaCl solution (50 ml)
14.61 g NaCl (MW: 58.44)
DDW to 50 ml - Modified low salt buffer (+ 0.04% SDS)
20 mM Tris, pH 8.0
2 mM EDTA
150 mM NaCl
0.1% Triton X-100
0.04% SDS
1x protease inhibitors
5 U/ml RNase inhibitor
For preparing 50 ml:
1 ml 1 M Tris (pH 8.0)
200 µl 0.5 M EDTA
1.5 ml 5 M NaCl
50 µl Triton X-100
200 µl 10% SDS (add before use)
5 tablets cOmplete-Mini (add before use)
6.25 µl RNasin plus (40 U/ml) (add before use)
47.05 ml DDW - TBS
50 mM Tris (pH 7.5)
150 mM NaCl
For preparing 10 ml:
500 µl 1 M Tris (pH 7.5)
300 µl 5 M NaCl
9.2 ml DDW - TBS-IGEPAL CA-630
50 mM Tris (pH 7.5)
150 mM NaCl
0.1% IGEPAL CA-630
For preparing 10 ml:
9.99 ml TBS
10 µl IGEPAL CA-630 - Elution buffer
50 mM Tris (pH 7.5)
150 mM NaCl
0.1% IGEPAL CA-630
500 µg/ml 3xFLAG peptide
For preparing 500 µl:
50 µl 5 mg/ml 3xFLAG peptide in TBS
450 µl TBS-IGEPAL CA-630
Acknowledgments
This protocol was adapted from the previously published paper (Fujita et al., 2017a). We thank M. Yuno for technical assistance. This work was supported by Grant-in-Aid for Scientific Research (C) (#15K06895) (T.F.) and Grant-in-Aid for Scientific Research (B) (#15H04329) (T.F., H.F.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
T.F. and H.F. have patents on enChIP (“Method for isolating specific genomic region using molecule binding specifically to endogenous DNA sequence”; patent number: Japan 5,954,808; patent application number: WO2014/125668). T.F. and H.F. are founders of Epigeneron, LLC.
References
- Fujita, T. and Fujii, H. (2014a). Efficient isolation of specific genomic regions retaining molecular interactions by the iChIP system using recombinant exogenous DNA-binding proteins. BMC Mol Biol 15: 26.
- Fujita, T. and Fujii, H. (2014b). Identification of proteins interacting with genomic regions of interest in vivo using engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP). Bio Protoc 4(10): e1124.
- Fujita, T. and Fujii, H. (2015). Isolation of specific genomic regions and identification of associated molecules by engineered DNA-binding molecule-mediated chromatin immunoprecipitation (enChIP) using CRISPR. Methods Mol Biol 1288: 43-52.
- Fujita, T. and Fujii, H. (2016a). Biochemical analysis of genome functions using locus-specific chromatin immunoprecipitation technologies. Gene Regul Syst Bio 10(Suppl 1): 1-9.
- Fujita, T., Kitaura, F., Yuno, M., Suzuki, Y., Sugano, S. and Fujii, H. (2017a). Locus-specific ChIP combined with NGS analysis reveals genomic regulatory regions that physically interact with the Pax5 promoter in a chicken B cell line. DNA Res 24(5): 537-548.
- Fujita, T., Yuno, M. and Fujii, H. (2016b). Efficient sequence-specific isolation of DNA fragments and chromatin by in vitro enChIP technology using recombinant CRISPR ribonucleoproteins. Genes Cells 21(4): 370-377.
- Fujita, T., Yuno, M., Suzuki, Y., Sugano, S. and Fujii, H. (2017b). Identification of physical interactions between genomic regions by enChIP-Seq. Genes Cells 22(6): 506-520.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Fujita, T. and Fujii, H. (2017). In vitro Engineered DNA-binding Molecule-mediated Chromatin Immunoprecipitation (in vitro enChIP) Using CRISPR Ribonucleoproteins in Combination with Next-generation Sequencing (in vitro enChIP-Seq) for the Identification of Chromosomal Interactions. Bio-protocol 7(22): e2612. DOI: 10.21769/BioProtoc.2612.
Category
Molecular Biology > DNA > DNA structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









