- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Preparation of the Partially Methylated Alditol Acetates Derived from CS Tetrasaccharides Containing Galactose for the Gas Chromatography/Mass Spectrometry Analysis
Published: Vol 7, Iss 21, Nov 5, 2017 DOI: 10.21769/BioProtoc.2600 Views: 10254
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Keratan Sulfate Disaccharide-branched Chondroitin Sulfate E from Mactra chinensis
Kyohei Higashi and Toshihiko Toida
Aug 5, 2017 7382 Views

Chromatographic Assays for the Enzymatic Degradation of Chitin
Sophanit Mekasha [...] Vincent G. H. Eijsink
May 5, 2021 5463 Views

Detailed Method for the Purification of Rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana
Liang Zhang [...] Breeanna R. Urbanowicz
Feb 5, 2026 144 Views
Abstract
Chondroitin sulfate (CS), a member of the glycosaminoglycan (GAG) family of carbohydrates, is composed of linear, sulfated repeating disaccharide sequences of N-acetyl-D-galactosamine (GalNAc) and glucuronic acid (GlcA). Recently, a keratan sulfate (KS) disaccharide [GlcNAc6S(β1-3)Galactose(β1-]-branched CS-E was identified from the clam species M. chinensis. This protocol details a methodology to analyze the glycosidic linkages of galactose in KS disaccharide-branched CS by GC-MS analysis. A complementary method for the identification and characterization of KS-branched CS in M. chinensis can be found in Higashi et al. (2016).
Keywords: Mactra chinensisBackground
Gas chromatography/mass spectrometry (GC-MS) analysis of the reaction products of partially methylated alditol acetates (PMAA) derived from the polysaccharides has been shown to represent a powerful tool to investigate the glycosidic linkages. The PMAAs preparation from M. chinensis in this protocol was performed according to the method of Anumula and Tayler (1992) with minor modifications.
Materials and Reagents
- Screw-cap tube (AGC techno glass, borosilicate Pyrex glass, 13 mm i.d. x 120 mm)
- Glass measuring pipette (0.1, 0.5, 1 and 2 ml)
- Pasteur pipette IK-PAS-5P (IWAKI, catalog number: 73-0001 )
- Keratan sulfate from Bovine Cornea (SEIKAGAKU, catalog number: 400760 )
- Dry tetrasaccharide (100 μg) is composed of ∆UA, N-acetyl-D-galactosamine (4S, 6S), galactose, N-acetyl-D-glucosamine (6S). Briefly, KS branched CS from M. chinensis was treated with chondroitinase ACII, and resulting tetrasaccharide was collected through the fractionation using HPLC with Docosil column. Please see details in Higashi et al. (2016)
- Dimethyl sulfoxide, dehydrated (dry DMSO) (Wako Pure Chemical Industries, catalog number: 040-18032 )
- Iodomethane (CH3I) (Wako Pure Chemical Industries, catalog number: 139-02662 )
- Chloroform
- Nitrogen gas (> 99.995%) (Nippon Megacare)
- Trifluoroacetic acid (TFA) (Wako Pure Chemical Industries, catalog number: 204-02743 )
- Acetic acid (NACALAI TESQUE, catalog number: 00212-43 )
- 4-N,N-dimethylaminopyridine (Wako Pure Chemical Industries, catalog number: 042-19212 )
- Pyridine (NACALAI TESQUE, catalog number: 29509-25 )
- Acetic anhydride (Wako Pure Chemical Industries, catalog number: 011-00276 )
- Hexane (NACALAI TESQUE, catalog number: 17935-05 )
- Sodium hydroxide (NaOH) (NACALAI TESQUE, catalog number: 31511-05 )
- Methanol (NACALAI TESQUE, catalog number: 21915-93 )
- Dimethyl sulfoxide (DMSO) (Wako Pure Chemical Industries, catalog number: 043-07216 )
- 0.5 mol/L hydrochloric acid methanolic solution (Wako Pure Chemical Industries, catalog number: 080-07725 )
- Sodium tetrahydroborate (NaBH4) (Wako Pure Chemical Industries, catalog number: 192-01472 )
- NaOH-DMSO suspension (see Recipes)
- 5% (v/v) pyridine in 50% acetonitrile/water (see Recipes)
- NaBH4 (5 mg/ml) solution (see Recipes)
Equipment
- Test tube mixer (SEIKAGAKU, model: TM-251 )
- Centrifuge (KUBOTA, model: Model 5922 )
- Sample concentrator (Hangzhou Allsheng Instruments, model: MD200-2 )
- Time-of-flight mass spectrometer JMS-T100GCV (JEOL, model: JMS-T100GCV )
- ZB-5ms column (0.25 µm film thickness, 0.25 µm i.d. x 30 m) (Phenomenex)
- Agilent Technologies 7890A GC system (Agilent Technologies, model: Agilent 7890A GC )
Procedure
The preparation of partially methylated alditol acetates (PMAAs) is depicted in Figure 1. All reagents are added using glass pipettes.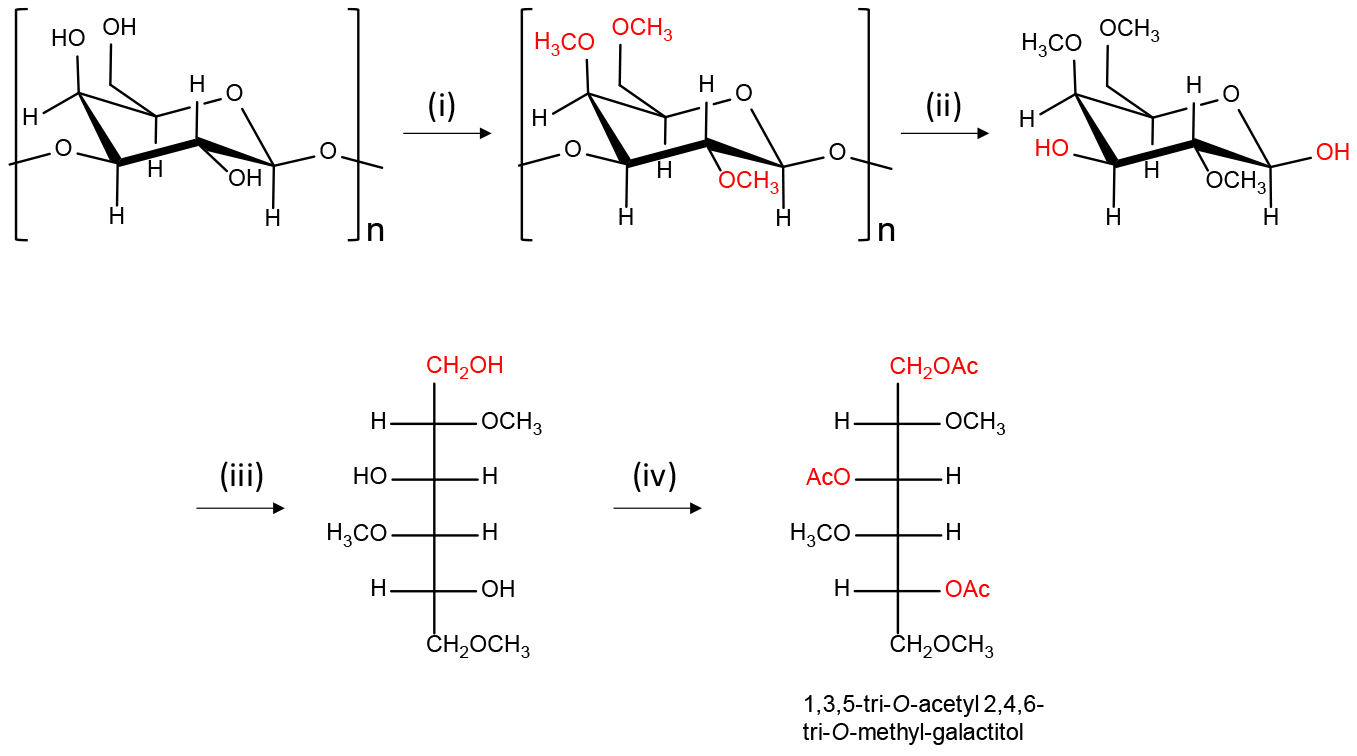
Figure 1. Scheme of the preparation of partially methylated alditol acetates (PMAAs) from polysaccharides having galactose residue. (i) DMSO, NaOH-DMSO suspension, CH3I; (ii) TFA (2.5 M); (iii) 5% pyridine in 50% CH3CN/H2O, NaBH4 in 30% CH3OH containing 0.03 M NaOH; (iv) 4-N,N-dimethylaminopyridine in CH3CN, pyridine, acetic anhydride.
- Preparation of partially methylated sugars
- Dry tetrasaccharide (100 μg) in a screw-cap tube.
- Add 200 μl of dry DMSO and sonicate for 30 min.
- Add 200 μl of NaOH-DMSO (see Recipes) and vortex.
- Transfer the tube containing the sample onto ice and add 100 μl of iodomethane (CH3I).
- Sonicate for 5 min and vortex 3 times.
- Add 50 μl of CH3I again, and incubate for 30 min on ice.
- Add 1 ml of distilled water and then vortex.
- Add 1 ml of chloroform using a glass pipet (1 ml), vortex for 1 min, shake the tube well for 1 min, vortex for 1 min.
- Centrifuge sample at 900 x g for 5 min at room temperature.
- Remove water fraction (upper layer) using Pasteur pipette (aspiration).
- To wash organic layer containing the methylated carbohydrates, add 1 ml of water and shake the tube well for 1 min, then vortex for 30 sec. Centrifuge at 900 x g for 5 min at room temperature and remove the water fraction using Pasteur pipette (aspiration).
- Wash the organic layer two times using 1 ml of water.
- Evaporate organic fraction (lower layer) for 40 min at 40 °C under a stream of nitrogen using a sample concentrator (Figure 2).
- Dissolve sample with 150 μl of 2.5 mol/L TFA. Fill with nitrogen gas and close tightly with a screw cap. Incubate at 100 °C for 4 h.
- Evaporate sample for 40 min at 40 °C under a stream of nitrogen using a sample concentrator (Figure 2).
- Add 500 μl of 5% pyridine in 50% acetonitrile/water (see Recipes) to the dried, partially methylated sugars.

Figure 2. Evaporation of sample at 40 °C under a stream of nitrogen using sample concentrator (MD200-2). The spray nozzle was inserted into a glass tube which was placed to aluminum dry bath heating block. Sample was dried at 40 °C under nitrogen gas.
- Dry tetrasaccharide (100 μg) in a screw-cap tube.
- Preparation of partially methylated alditol acetates (PMAAs) from partially methylated sugars
- Add 200 μl of 5 mg/ml of NaBH4 solution (see Recipes) into the sample and incubate for 4 h at 37 °C.
- Add 200 μl of acetic acid and evaporate sample within 1 h at 40 °C under a stream of nitrogen using a sample concentrator (Figure 2).
- Dissolve sample with 1 ml of 0.1% (v/v) MeOH-HCl and evaporate sample within 1 h at 40 °C under a stream of nitrogen (Figure 2). Repeat this step four times.
- Add 150 μl of 5 mg/ml of 4-N,N-dimethylaminopyridine, 50 μl of pyridine and 150 μl of acetic anhydride, respectively, and incubate at room temperature for 4 h.
- After incubation, add 2 ml of distilled water.
- Add 2 ml of chloroform, vortex, centrifuge at 900 x g for 5 min at room temperature and remove water (upper layer).
- Add 2 ml of distilled water, vortex, centrifuge at 900 x g for 5 min and remove water (upper layer).
- Evaporate sample (organic layer) for 40 min at 40 °C under a stream of nitrogen (Figure 2).
- Dissolve sample with 30 μl of hexane.
- Add 200 μl of 5 mg/ml of NaBH4 solution (see Recipes) into the sample and incubate for 4 h at 37 °C.
- GC-MS (Agilent Technologies 7890A GC system) analysis of PMAAs
- Set electro impact ionization, 70 eV.
- Set carrier gas, helium at 1.2 ml/min.
- Set split less sample injection.
- Set column oven temperature program: 3 min at 100 °C, with an increase at 4 °C/min to 160 °C, 1 min at 160 °C followed by an increase at 0.5 °C/min to 180 °C, and a final increase at 20 °C/min to 260 °C and held for 10 min at 260 °C.
- Submit 1 μl of PMAAs in hexane to GC-MS.
- Set electro impact ionization, 70 eV.
Data analysis
Electron ionization (EI) which afford the fragment-masses of small compounds is the most common form of ionization for GC-MS analysis. In general, fragmentation patterns generated by EI are compound dependent. In case of the structural analysis of glycans, electron ionization chromatogram (EIC) of certain fragment ions (m/z 45, 117, 161, 233) are useful for identifying peaks corresponding to PMAAs (Björndal et al., 1970). When EIC of m/z 233 was monitored, PMAA (1,3,5-tri-O-acetyl 2,4,6-tri-O-methyl-galactitol) from galactose in tetrasaccharide of M. chinensis was detected at 28.68 min (Higashi et al., 2016). However, PMAAs from ∆UA, N-acetyl-D-galactosamine and N-acetyl-D-glucosamine were not observed.
Notes
- For isolation of KS branched CS-E from M. chinensis and preparation of tetrasaccharide from KS branched CS-E, please see details in Higashi et al. (2016). In general, quantitation of CS was performed by post-column HPLC through the detection of unsaturated disaccharides obtained by chondroitinase (Chase). Interestingly, unknown peaks were found when CS from M. chinensis was treated with Chase ACII (at high concentrations) but not Chase ABC. Thus, unknown peaks were collected and analyzed by LC-MS/MS and GC-MS. In LC-MS/MS analysis, we found that unknown peaks consisted of tetrasaccharides including ∆UA, N-acetyl-D-galactosamine (4S, 6S), hexose, HexNAc (S). In addition, GC-MS analysis suggested that hexose in tetrasaccharide was galactose. Finally, N-acetyl-D-glucosamine (6S) in tetrasaccharide was suggested by 2D-NMR.
- PMAAs from M. chinensis were prepared according to the method of Anumula and Taylor (1992) with minor modifications.
Recipes
- NaOH-DMSO suspension (prior preparation)
- Combine 0.2 ml of 50% (w/w %) NaOH and 0.4 ml of methanol in a screw-cap tube
- Dilute with 6 ml of DMSO, vortex and sonicate for 3-5 min
- Centrifuge the fine dispersion of NaOH in DMSO at 900 x g for 10 min at room temperature
- Resuspend obtained precipitate with 6 ml of fresh DMSO and centrifuge at 900 x g for 10 min at room temperature. Repeat this step two times
- Resuspend obtained precipitate with 6 ml of dry DMSO, dehydrated and centrifuge at 900 x g for 10 min at room temperature. Repeat this step three times
- Suspend precipitate with 4 ml of dry DMSO and store at 4 °C
- Combine 0.2 ml of 50% (w/w %) NaOH and 0.4 ml of methanol in a screw-cap tube
- 5% (v/v) pyridine in 50% acetonitrile/water (prior preparation)
- Prepare 1.9 ml of 50% acetonitrile with distilled water
- Combine 0.1 ml of pyridine and 1.9 ml of 50% acetonitrile/water
- Prepare 1.9 ml of 50% acetonitrile with distilled water
- NaBH4 (5 mg/ml) in 30% methanol containing 0.03 mol/L of NaOH (prior preparation)
- Prepare a solution of 0.03 mol/L NaOH in 30% MeOH
- Dissolve NaBH4 with 30% MeOH containing 0.03 mol/L of NaOH
- Prepare a solution of 0.03 mol/L NaOH in 30% MeOH
Acknowledgments
We thank Dr. Sayaka Masuko for her help in preparing this manuscript. This protocol was adapted from the original work (Higashi et al., 2016) to provide the detailed procedures. This study was supported in part by the Grant-in-aid for Scientific Research from the Ministry of Education, Culture, Sport, Science and Technology of Japan and the Inohana Foundation, Chiba University. The authors declare that there are no conflicts of interest.
References
- Anumula, K. R. and Taylor, P. B. (1992). A comprehensive procedure for preparation of partially methylated alditol acetates from glycoprotein carbohydrates. Anal Biochem 203(1): 101-108.
- Björndal, H., Hellerqvist, C. G., Lindberg, B. and Svensson, S. (1970). Gas‐liquid chromatography and mass spectrometry in methylation analysis of polysaccharides. Angew Chem Int Ed Engl 9: 610-619.
- Higashi, K., Takeda, K., Mukuno, A., Okamoto, Y., Masuko, S., Linhardt, R. J. and Toida, T. (2016). Identification of keratan sulfate disaccharide at C-3 position of glucuronate of chondroitin sulfate from Mactra chinensis. Biochem J 473(22): 4145-4158.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Higashi, K. and Toida, T. (2017). Preparation of the Partially Methylated Alditol Acetates Derived from CS Tetrasaccharides Containing Galactose for the Gas Chromatography/Mass Spectrometry Analysis. Bio-protocol 7(21): e2600. DOI: 10.21769/BioProtoc.2600.
Category
Biochemistry > Carbohydrate > Polysaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









