- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Ensifer-mediated Arabidopsis thaliana Root Transformation (E-ART): A Protocol to Analyse the Factors that Support Ensifer-mediated Transformation (EMT) of Plant Cells
Published: Vol 7, Iss 19, Oct 5, 2017 DOI: 10.21769/BioProtoc.2564 Views: 11805
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Efficient Genetic Transformation and Suicide Plasmid-mediated Genome Editing System for Non-model Microorganism Erwinia persicina
Tingfeng Cheng [...] Lei Zhao
Mar 20, 2024 2442 Views

Purification of Native Dentilisin Complex from Treponema denticola by Preparative Continuous Polyacrylamide Gel Electrophoresis and Functional Analysis by Gelatin Zymography
Pachiyappan Kamarajan [...] Yvonne L. Kapila
Apr 5, 2024 2095 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1671 Views
Abstract
Ensifer adhaerens OV14, a soil borne alpha-proteobacteria of the Rhizobiaceae family, fortifies the novel plant transformation technology platform termed ‘Ensifer-mediated transformation’ (EMT). EMT can stably transform both monocot and dicot species, and the host range of EMT is continuously expanding across a diverse range of crop species. In this protocol, we adapted a previously published account that describes the use of Arabidopsis thaliana roots to investigate the interaction of A. thaliana and Agrobacterium tumefaciens. In our laboratory, we routinely use A. thaliana root explants to examine the factors that enhance the utility of EMT. In addition, the E-ART protocol can be used to study the transcriptional response of E. adhaerens and host plant following exposure to explant tissue, the transformability of different Ensifer adhaerens strains/mutants as well as testing the susceptibility of A. thaliana mutant lines as a means to decipher the mechanisms underpinning EMT.
Keywords: Ensifer adhaerens strain OV14Background
The advancement of Ensifer-mediated transformation (EMT) technology to successfully transform dicots viz., Arabidopsis thaliana, Solanum tuberosum, Nicotiana tabacum, Manihot esculenta, Brassica napus, and the monocot; Oryza sativa, has been previously reported (Wendt et al., 2012; Zuniga-Soto et al., 2015; Chavarriaga-Aguirre et al., 2016; Rathore et al., 2016). Additionally, genomic analysis of E. adhaerens OV14 by Rudder et al. (2014) revealed the bacterium possessed a 7.7 Mb genome comprised of two circular chromosomes (3.96 Mb and 2.01 Mb) and two plasmids (1.61 Mb and 125 Kb). A comparative analysis of the genome of E. adhaerens OV14 with Agrobacterium tumefaciens C58 (classic genetic engineer) and Sinorhizobium meliloti 1021 (a rhizobia with a propensity for low rates of genetic transformation; Broothaerts et al., 2005), highlighted that both E. adhaerens OV14 and S. meliloti 1021 contain homologs to several chromosomal-based genes essential for Agrobacterium-mediated transformation (AMT). However, a suite of genes which positively influence the successful transformation of a plant genome were found to be only present in the genome of E. adhaerens OV14 but absent from S. meliloti 1021 (Rudder et al., 2014). Overall, the sequence analysis of the E. adhaerens OV14 genome has significantly expanded the knowledge base describing the genetic systems that regulate the transformation of plant genomes via EMT.
To date several transient transformation systems have been proposed to study gene function in plants in regard to the use of A. tumefaciens (Wroblewski et al., 2005; Gelvin, 2006; Bhaskar et al., 2009; Li et al., 2009; Van Loock et al., 2010; Hwang et al., 2013; Krenek et al., 2015). Whereas, the stable transformations of any plant species are lengthy processes to test the utility of a bacterial transfection system to transform the plants. The advantages of transient transformation methods include rapid production of results, functional genomic studies and recombinant protein production (Van Loock et al., 2010; Krenek et al., 2015). The model plant A. thaliana is a powerful research tool with which to study the molecular, genetic and biochemical processes that support genetic transformation of somatic tissues as well as in planta (Provart et al., 2016). In contrast to the several months typically required for the transformation of primary crop species, these investigations require a rapid, reproducible and easy quantification method to determine the rate of transient transformation. Previously, Gelvin (2006) reported an efficient and reproducible quantitative assay for Agrobacterium-mediated Transformation using A. thaliana roots. This assay has well served the purpose of testing either Agrobacterium strains or A. thaliana ecotypes/mutants in the authors’ laboratory for more than two decades, reflecting the significance and competency of the assay in publications such as Shi et al. (2014). Conversely, a transient transformation method to facilitate comparable studies via EMT is not available. In response, the Ensifer-mediated A. thaliana Root Transformation (E-ART) protocol presented here is designed to address this deficit so that specific genetic and microbiological factors that support/enhance EMT can be identified to support the application of the technology to agronomically important crop species. The protocol is a modified version of an existing AMT based quantitative A. thaliana roots assay (Gelvin, 2006). While developing the E-ART protocol, we learnt that it is possible to improve transient GUS expression in A. thaliana root segments by adjusting several experimental factors (e.g., time of co-cultivation, acetosyringone concentrations, etc.) involved in the early stages of E. adhaerens transfection. E-ART, being the first quantitative method of transient gene expression for EMT will facilitate the rapid evaluation of novel E. adhaerens strains in plant transformation while also providing a platform to assess the genetic response of plants to EMT.
Materials and Reagents
- 2 ml centrifuge tubes
- Square Petri-dishes (Greiner Bio One International, catalog number: 688161 )
- Parafilm (Bemis, catalog number: PM992 )
- 50 ml Falcon tube
- Scalpel blades (NO. 10A, Swan Morton, catalog number: 0302 )
- Sterile filter papers (GE Healthcare, catalog number: 1004-090 )
- Petri dish 92 x 16 mm w/o cams (SARSTEDT, catalog number: 82.1472 )
- A. thaliana seed (in this case ecotype Columbia, Col-0)
Note: A. thaliana seed is no more than 6 months old being stored at 4 °C.
- E. adhaerens strain OV14 harbouring plasmid of choice (in this case pCambia5105/pCambia5106 plasmids [Jefferson et al., 2006])
- 70% ethanol
- Distilled sterile water (DSW)
- Bleach (5% sodium hypochlorite; final concentration used is 50% Bleach, i.e., 1:1 Bleach:water)
- Tween-20
- Agarose (0.1%, Sigma-Aldrich, catalog number: A9539-500G )
- Antibiotics for bacterial selection: kanamycin, streptomycin, spectinomycin (Duchefa Biochemie)
- Sodium chloride (0.9% NaCl solution)
- Acetosyringone (Sigma-Aldrich, catalog number: D134406 )
- Cefotaxime sodium (Duchefa Biochemie, catalog number: C0111.0005 )
- Tryptone (Oxoid, catalog number: LP0042 )
- Yeast extract (Oxoid, catalog number: LP0021 )
- Calcium chloride dehydrate (Duchefa Biochemie, catalog number: C0504 )
- Agar No. 1 (Oxoid, catalog number: LP0011 )
- MS basal salts (Duchefa Biochemie, catalog number: M0221 )
- Sucrose (Duchefa Biochemie, catalog number: S0809 )
- 2,4-Morpholino-ethane sulfonic acid (MES monohydrate) (Duchefa Biochemie, catalog number: M1503 )
- Myo-inositol (Duchefa Biochemie, catalog number: I0609 )
- Nicotinic acid (Duchefa Biochemie, catalog number: N0611 )
- Pyridoxine (Duchefa Biochemie, catalog number: P0612 )
- Thiamine-HCl (Duchefa Biochemie, catalog number: T0614 )
- D-Glucose monohydrate (Duchefa Biochemie, catalog number: G0802 )
- Indole-3-acetic acid (IAA) (Duchefa Biochemie, catalog number: I0901 )
- 2,4-Dicholorophenocxyacetic acid (2,4-D) (Duchefa Biochemie, catalog number: D0911 )
- Kinetin (Duchefa Biochemie, catalog number: K0905 )
- X-GlcA cyclohexylammonium salt (Duchefa Biochemie, catalog number: X1405 )
- Dimethyl sulfoxide (DMSO) (Duchefa Biochemie, catalog number: D1370 )
- Teagasc-Tryptone Yeast-extract (TTY) medium (Rathore et al., 2015) (see Recipes)
- MS based media (see Recipes)
- Seed germination media (SGM)
- Co-cultivation media (CCM)
- Callus Induction media (CIM)
- Vitamin stock
- Seed germination media (SGM)
- X-GlcA solution (see Recipes)
- Histochemical GUS stain solution (Jefferson et al., 1987) (see Recipes)
Equipment
- Pipettes (P1000, P100, P10)
- Controlled environment room/chamber to grow healthy A. thaliana plants (24 °C, 16 h light, 8 h dark), abbreviated as CT room
- Conical flasks (250 ml, sterile)
- Centrifuge
- Incubator (28 °C and 37 °C)
- Shaker incubator (28 °C, 220 rpm)
- Fridge (4 °C) and freezers (-20 °C and -80 °C)
- Laminar flow to perform aseptic work
- Bead sterilizer/flame to sterilize forceps
- Scalpel blades handles (No.7 S/S, Swan Morton, catalog number: 0907 )
- Autoclave (15 min at 121 °C and 15 psi)
- pH meter
- Weighing balance
- NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM 2000 )
- Elga water purifier (Veolia Water Solution & Technologies, model: PURELAB® OPTION-R 7, catalog number: OR007BPM1 )
- Stereo-microscope
Software
- SAS system (Version 9.3, copyright 2002-2010 by SAS Institute Inc., Cary, NC, USA)
Procedure
- Seed sterilization and germination
- Surface sterilize A. thaliana seeds (~5 mg) in a 2 ml tube with 70% ethanol (EtOH) by vortexing the tube for 2 min. Discard the EtOH with an adjustable micropipette (1,000 µl) and wash seeds twice with DSW. All the steps were performed under a laminar flow hood to ensure aseptic conditions throughout.
- Add 50% Bleach containing 0.1% Tween-20 to the seeds and sterilize for 10 min with vigorous shaking. Once complete, decant Bleach with a 1,000 µl micropipette (Figure 1A).
- Rinse the seeds for five times with DSW to remove any residue of Bleach.
- Post-sterilization process, add 0.1% agarose (previously autoclaved for 15 min at 121 °C) to seeds for ease of handling.
- Using a micropipette (1,000 µl), place ~4 seeds per drop aligned in a row leaving ~1 inch distance from the top end of square Petri-dishes (Figures 1B and 1C) containing MS based seed germination medium (SGM, see Recipes). Double wrap the Petri-dishes using Parafilm.
- Incubate the Petri-dishes vertically in a stand (to ensure elongated root formation) in a CT (controlled temperature) room set at 24 °C, 16 h light ~4,000 lux photoperiod and 8 h dark for 15-20 days (Figure 1D).
- Root harvesting should be achieved before roots reach the end of dish, at which stage ~20 segments of < 0.5 cm length each should be obtained per plant.

Figure 1. Arabidopsis thaliana seed sterilisation and germination. A. The A. thaliana ecotype Col-0 in 50% Bleach in a 2 ml centrifuge tube; B. Placing sterilised seeds on square Petri-dishes leaving ~1 inch top space for plants to grow; C. Plated seeds on SGM; D. Vertically aligned Petri-dishes in CT room (at 24 °C, 16 h photoperiod) showing plantlets with roots.
- Surface sterilize A. thaliana seeds (~5 mg) in a 2 ml tube with 70% ethanol (EtOH) by vortexing the tube for 2 min. Discard the EtOH with an adjustable micropipette (1,000 µl) and wash seeds twice with DSW. All the steps were performed under a laminar flow hood to ensure aseptic conditions throughout.
- Preparation of Ensifer inoculums
- Grow E. adhaerens OV14_pCambia5105 (here onwards E5105) on a TTY agar (see Recipes) containing 100 mg/L kanamycin, 200 mg/L streptomycin and 200 mg/L spectinomycin to select for the unitary plasmid containing T-DNA as well as virulence (vir) genes, overnight at 28 °C (Figure 2A).
- Pick a single colony to inoculate 10 ml TTY broth containing appropriate antibiotics (100 mg/L kanamycin, 200 mg/L streptomycin and spectinomycin) in a sterile conical flask and grow overnight at 28 °C and 220 rpm to reach 0.8 OD600 nm (Figures 2B and 2C).
- Pellet the cells in 50 ml Falcon tubes by centrifugation and wash them using 0.9% NaCl to remove any antibiotic residues. Centrifuge bacterial cultures at 3,750 x g (rcf) for 25-30 min for the inoculum preparation. Re-suspend the cells in 0.9% NaCl to maintain 0.8 OD600 nm (Figure 2D).
- Add 200 µM acetosyringone and incubate at 28 °C, 220 rpm for additional 1.5 h to induce virulence.
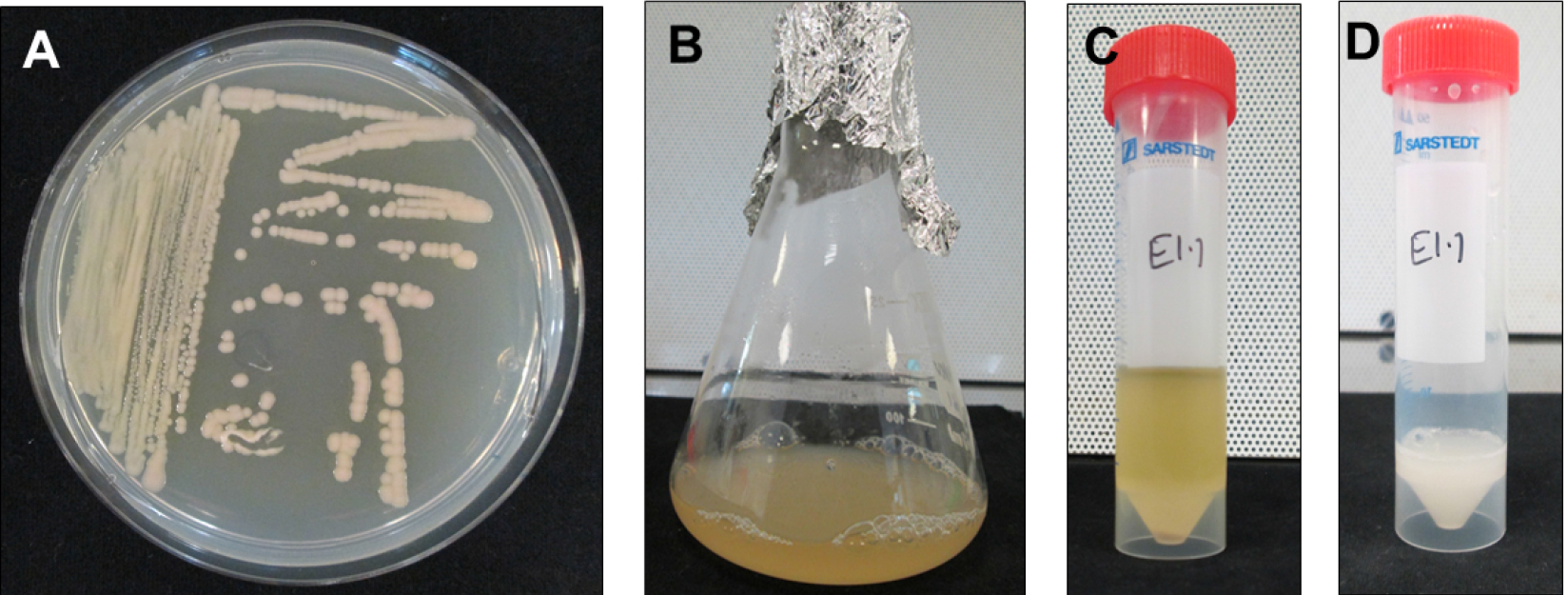
Figure 2. Steps involved in preparing E5105 cultures for transfection of root segments. A. Overnight grown E5105 on TTY agar containing 100 mg/L kanamycin + 200 mg/L streptomycin + 200 mg/L spectinomycin to obtain single colonies. B. Single colony derived E5105 culture in TTY broth with appropriate antibiotics. C. E5105 culture in a 50 ml Falcon tube for centrifugation to remove TTY broth + antibiotics. D. E5105 resuspended in 0.9% NaCl and supplemented with 200 µM acetosyringone for vir induction at 28 °C, 1.5 h.
- Grow E. adhaerens OV14_pCambia5105 (here onwards E5105) on a TTY agar (see Recipes) containing 100 mg/L kanamycin, 200 mg/L streptomycin and 200 mg/L spectinomycin to select for the unitary plasmid containing T-DNA as well as virulence (vir) genes, overnight at 28 °C (Figure 2A).
- Preparation and transfection of A. thaliana root segments
- Separate the A. thaliana roots from plants using a sterile scalpel blade before laying roots down on a Petri-dish containing a small amount (~500 µl) of DSW to ensure adequate root hydration (Figures 3A and 3B).
- Align roots together and cut into 0.3 to 0.5 cm long segments (Figure 3C). Place bundles of ~30 root segments on MS based co-cultivation medium (CCM, see Recipes) containing 200 µM acetosyringone.
- Using a P1000 pipette place 2-3 drops of E5105 on each the A. thaliana root bundle and allow a transfection time of 10 min (Figure 3D). Untreated root segments (no bacteria) should be used as a negative control.
- Remove any excess bacterial solution after the 10 min transfection time using the P1000 pipette. Double wrap the Petri-dishes with Parafilm.
- Co-culture the bacteria and A. thaliana roots at 20 °C, in the dark for 5 days.
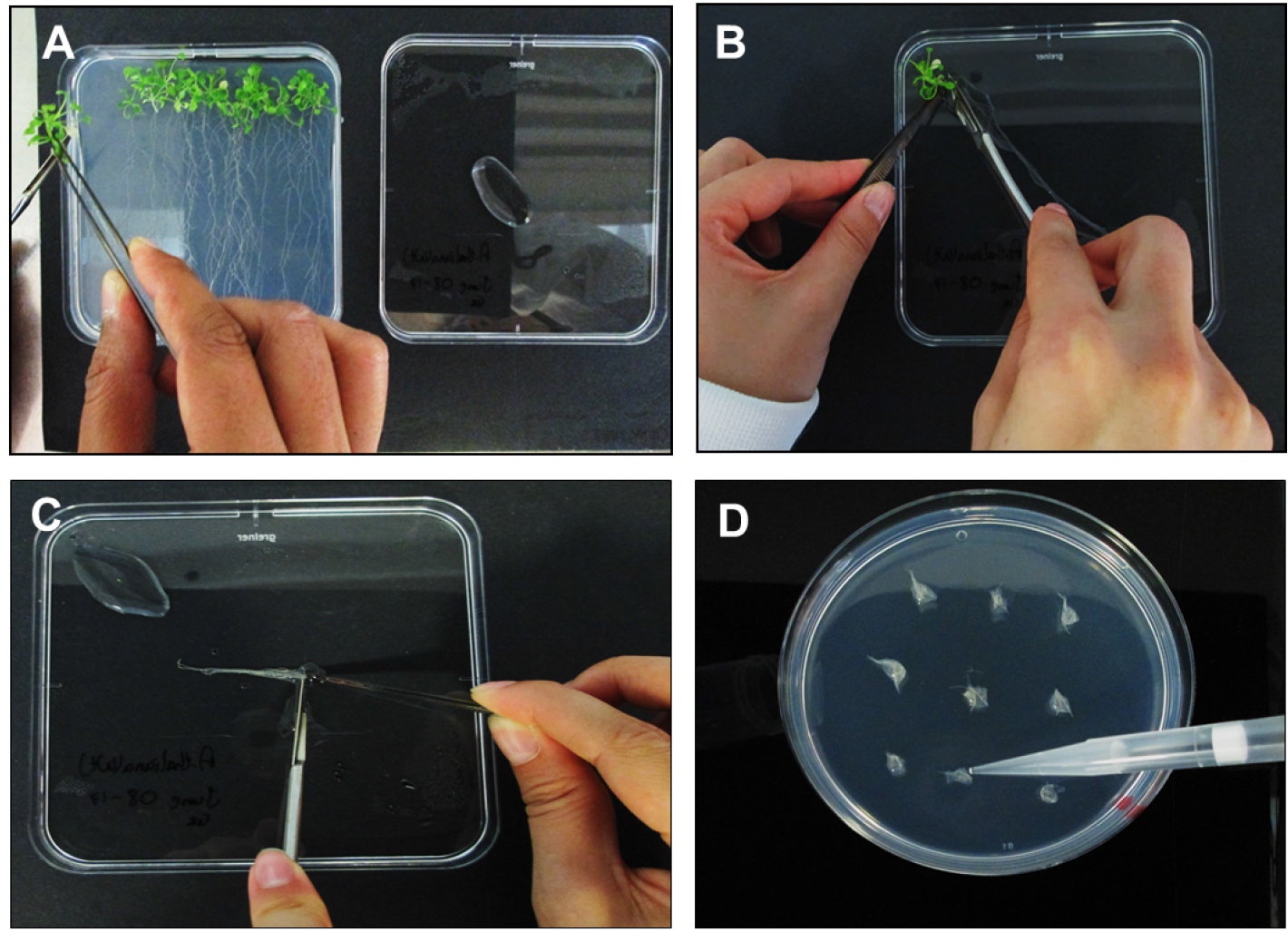
Figure 3. Processing of Arabidopsis thaliana plants and inoculations. A. Moving A. thaliana plants from SGM to square Petri-dish lid containing DSW; B. Separating the roots from the shoots; C. Cutting A. thaliana roots into ~3 mm segments; D. Inoculating the root segments with E5105 post 1.5 h vir induction.
- Separate the A. thaliana roots from plants using a sterile scalpel blade before laying roots down on a Petri-dish containing a small amount (~500 µl) of DSW to ensure adequate root hydration (Figures 3A and 3B).
- Transient GUS expression assay
- Post 5 days co-cultivation; wash treated/untreated root segments in DSW containing 500 mg/L cefotaxime to mitigate bacterial overgrowth.
- GUS staining: The activity of β-glucuronidase in putative transgenic root segments is performed as described by Jefferson et al. (1987) using one of two ways:
- For faster results, the washed roots are treated in GUS stain solution (see Recipes) with an overnight incubation at 37 °C. To remove the excess stain, wash root segments in 90% (v/v) ethanol (Figures 4A and 4B).
- To allow further proliferation of root segments and to facilitate ease of handling, transfer the roots onto callus induction medium (CIM, see Recipes) containing 250 mg/L cefotaxime. Seal the plates with Parafilm and incubate for additional 5-6 days at 24 °C in the dark, before staining in GUS stain solution, overnight at 37 °C (Figures 4C and 4D).
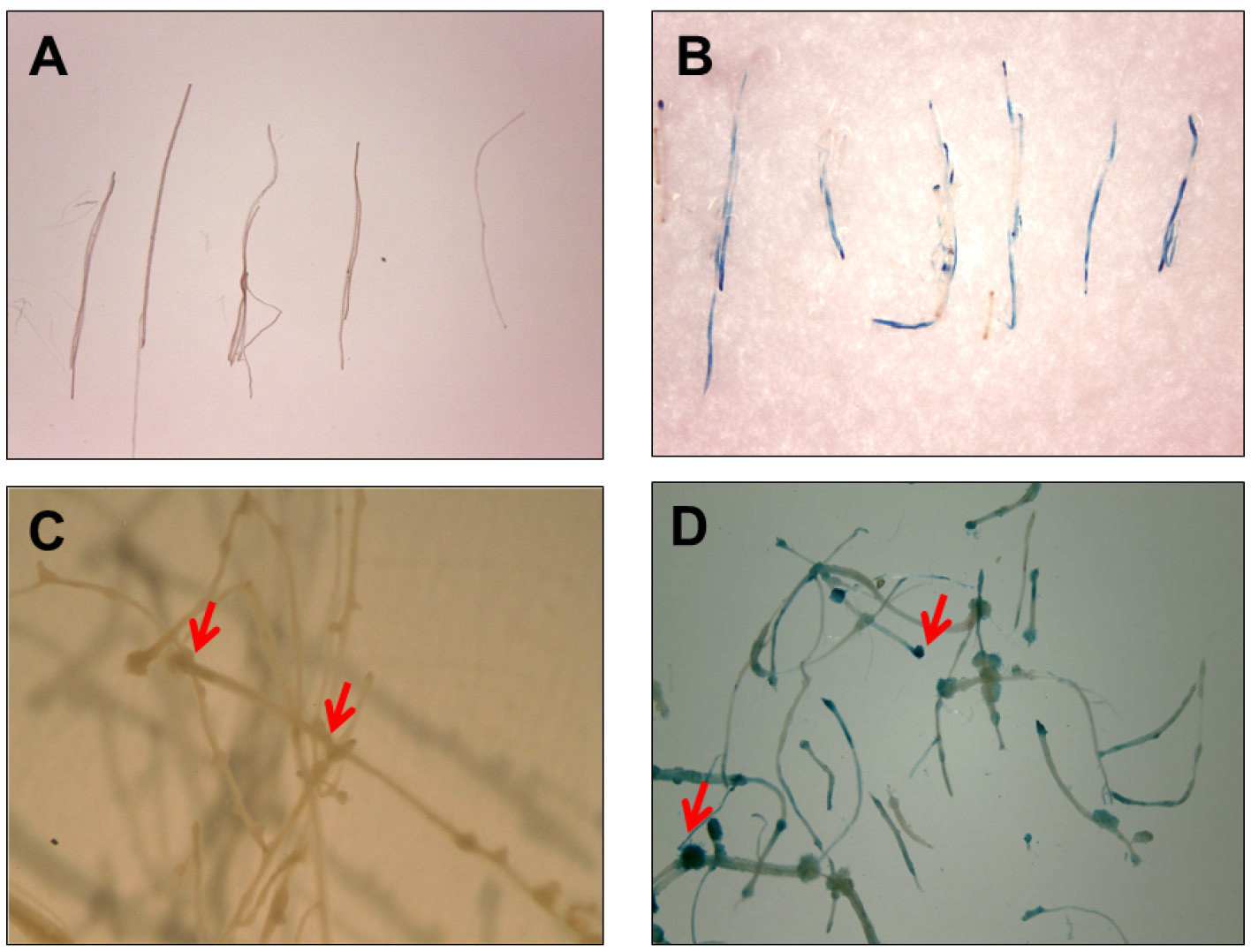
Figure 4. Histochemical GUS activity via E-ART. Root segments were immediately transferred to GUS stain post 5 days co-cultivation (A) untreated control and (B) treated with E5105. GUS staining of root segments showing callus formation (indicated with red arrows) when segments were transferred to CIM containing 250 mg/L cefotaxime for 6 days post co-cultivation (C) untreated control and (D) treated with E5105.
- For faster results, the washed roots are treated in GUS stain solution (see Recipes) with an overnight incubation at 37 °C. To remove the excess stain, wash root segments in 90% (v/v) ethanol (Figures 4A and 4B).
- Post 5 days co-cultivation; wash treated/untreated root segments in DSW containing 500 mg/L cefotaxime to mitigate bacterial overgrowth.
Data analysis
Perform all experiments in triplicate with three technical replicates (one Petri-dish containing 9 bundles of root explants was treated as one technical replicate). The transient expression of GUS in A. thaliana roots treated with E5105 (E. adhaerens OV14 carrying pCAMBIA5105) is measured based on GUS activity i.e., blue colouration. Count blue roots/spots containing root segments for each treatment in each replicate under a stereo-microscope and record in MS Excel (Microsoft, USA). For statistical significance, analyse data using the General Linear Model (GLM) in the SAS system (Version 9.3, copyright 2002-2010 by SAS Institute Inc., Cary, NC, USA). The GLM procedure enables t-test (LSD) and Tukey’s studentized Range (HSD) test using the SAS system.
Notes
- Store seeds in Falcon tubes, properly labelled for the ecotype at 4 °C.
- All steps are strictly performed under aseptic conditions in a laminar flow hood. All media are autoclaved (15 min at 121 °C) and media supplements such as growth hormones, antibiotics, are filter sterilised and added after autoclaving.
- Seal the Petri-dishes with a double layer of Parafilm to maintain humidity to grow healthy plants. It is primarily important for high transient GUS expression that the plants are young to provide healthy roots quickly.
- Always process the A. thaliana plants for obtaining root explants before a flower bolt emerges as older plants will lead to lower transformation rates as detailed by Gelvin (2006).
- More importantly, always use fresh bacterial cultures to obtain higher rates of transformations. Do not allow bacterial cultures (E5105) to grow over 0.8 OD600 nm. Higher OD cultures will overgrow bacteria during co-cultivation that will negatively influence the GUS expression.
- The transfection of roots should be performed within 30 min of cutting the root segments. Leaving cut root segments for a longer time will affect the transfection and transformation negatively.
- Always include untreated root explants (no bacterial inoculum) as a negative control in the experiment.
- Current work shows that a combination of 5 days co-cultivation time and 200 µM acetosyringone improved rate of transient GUS expression in the A. thaliana root segments.
Recipes
Note: The following media are prepared, autoclaved and stored at room temperature.
- Teagasc-tryptone yeast extract (TTY) medium
1.0% tryptone
0.5% yeast extract
After autoclaving add 20 ml of 1 M sterile CaCl2 per litre of medium
To make TTY agar, add 1.5% agar to TTY broth prior to autoclaving
- MS based media (Table 1)
Note: The MS based media are poured in Petri-dishes and can be stored at 4 °C for up to 15 days. Prepare vitamin stock, filter sterilise and store aliquots of 1 ml at -20 °C for up to 1 month.
Table 1. Media recipe used for Ensifer-mediated Arabidopsis thaliana root transformation (E-ART)

- X-GlcA solution
To prepare 1 M of X-GlcA solution:
Add 521.8 mg of X-GlcA to 10 ml DMSO and vortex to dissolve
Aliquot 1 ml in centrifuge tubes to store at -20 °C for long-term use
Note: This substance is hygroscopic and light sensitive.
- Histochemical GUS stain solution
Prepare GUS solution by adding below listed chemicals to reach the final concentrations as:
50 mM sodium phosphate buffer pH 7.0
1 mM EDTA
0.1 mM of ferri- and ferro-cyanide, individually
0.1% SDS
40 mM X-GlcA
Store aliquots at -20 °C
Acknowledgments
This research work was supported by the Teagasc Walsh Fellowship Scheme which funded DSR. Authors gratefully acknowledge Prof. Stanton B. Gelvin for the Agrobacterium transformation of Arabidopsis thaliana Roots: A Quantitative Assay (2006) published as a chapter in Methods in Molecular Biology, Agrobacterium protocols, which was adapted to develop E-ART protocol.
References
- Bhaskar, P. B., Venkateshwaran, M., Wu, L., Ane, J. M. and Jiang, J. (2009). Agrobacterium-mediated transient gene expression and silencing: a rapid tool for functional gene assay in potato. PLoS One 4(6): e5812.
- Broothaerts, W., Mitchell, H. J., Weir, B., Kaines, S., Smith, L. M., Yang, W., Mayer, J. E., Roa-Rodriguez, C. and Jefferson, R. A. (2005). Gene transfer to plants by diverse species of bacteria. Nature 433(7026): 629-633.
- Chavarriaga-Aguirre, P., Brand, A., Medina, A., Prias, M., Escobar, R., Martinez, J., Diaz, P., Lopez, C., Roca, W. M. and Tohme, J. (2016). The potential of using biotechnology to improve cassava: a review. In Vitro Cell Dev Biol Plant 52(5): 461-478.
- Gelvin, S. B. (2006). Agrobacterium transformation of Arabidopsis thaliana roots: a quantitative assay. Methods Mol Biol 343: 105-113.
- Hwang, H. H., Wu, E. T., Liu, S. Y., Chang, S. C., Tzeng, K. C. and Kado, C. I. (2013). Characterization and host range of five tumorigenic Agrobacterium tumefaciens strains and possible application in plant transient transformation assays. Plant Pathology 62: 1384-1397.
- Jefferson, R. A., Jefferson, O. A., Smith, L., Baillie, B. K., Raines, S., Ulkir, B., Tassie, A. and Tian, L. (2006). Freedom to co-operate: Transbacter as a Biological Open Source (BIOS) Tool for Gene Transfer. 8th international congress of plant molecular biology abstracts. Plant Mol Biol Rep 24: 141-160.
- Jefferson, R. A., Kavanagh, T. A. and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6(13): 3901-3907.
- Krenek, P., Samajova, O., Luptovciak, I., Doskocilova, A., Komis, G. and Samaj, J. (2015). Transient plant transformation mediated by Agrobacterium tumefaciens: Principles, methods and applications. Biotechnol Adv 33(6 Pt 2): 1024-1042.
- Li, J. F., Park, E., von Arnim, A. G. and Nebenfuhr, A. (2009). The FAST technique: a simplified Agrobacterium-based transformation method for transient gene expression analysis in seedlings of Arabidopsis and other plant species. Plant Methods 5: 6.
- Provart, N. J., Alonso, J., Assmann, S. M., Bergmann, D., Brady, S. M., Brkljacic, J., Browse, J., Chapple, C., Colot, V., Cutler, S., Dangl, J., Ehrhardt, D., Friesner, J. D., Frommer, W. B., Grotewold, E., Meyerowitz, E., Nemhauser, J., Nordborg, M., Pikaard, C., Shanklin, J., Somerville, C., Stitt, M., Torii, K. U., Waese, J., Wagner, D. and McCourt, P. (2016). 50 years of Arabidopsis research: highlights and future directions. New Phytol 209(3): 921-944.
- Rathore, D. S, Doohan, F. and Mullins, E. (2016). Capability of the plant-associated bacterium, Ensifer adhaerens strain OV14, to genetically transform its original host Brassica napus. Plant Cell Tiss Organ Cult 127: 85-94.
- Rathore, D. S., Lopez-Vernaza, M. A., Doohan, F., Connell, D. O., Lloyd, A. and Mullins, E. (2015). Profiling antibiotic resistance and electrotransformation potential of Ensifer adhaerens OV14; a non-Agrobacterium species capable of efficient rates of plant transformation. FEMS Microbiol Lett 362(17): fnv126.
- Rudder, S., Doohan, F., Creevey, C. J., Wendt, T. and Mullins, E. (2014). Genome sequence of Ensifer adhaerens OV14 provides insights into its ability as a novel vector for the genetic transformation of plant genomes. BMC Genomics 15: 268.
- Shi, Y., Lee, L. Y. and Gelvin, S. B. (2014). Is VIP1 important for Agrobacterium-mediated transformation? Plant J 79: 848-860.
- Van Loock, B., Markakis, M. N., Verbelen, J. P. and Vissenberg, K. (2010). High-throughput transient transformation of Arabidopsis roots enables systematic colocalization analysis of GFP-tagged proteins. Plant Signal Behav 5(3): 261-263.
- Wendt, T., Doohan, F. and Mullins, E. (2012). Production of Phytophthora infestans-resistant potato (Solanum tuberosum) utilising Ensifer adhaerens OV14. Transgenic Res 21(3): 567-578.
- Wroblewski, T., Tomczak, A. and Michelmore, R. (2005). Optimization of Agrobacterium-mediated transient assays of gene expression in lettuce, tomato and Arabidopsis. Plant Biotechnol J 3(2): 259-273.
- Zuniga-Soto, E., Mullins, E. and Dedicova, B. (2015). Ensifer-mediated transformation: an efficient non-Agrobacterium protocol for the genetic modification of rice. Springerplus 4: 600.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rathore, D. S., Doohan, F. M. and Mullins, E. (2017). Ensifer-mediated Arabidopsis thaliana Root Transformation (E-ART): A Protocol to Analyse the Factors that Support Ensifer-mediated Transformation (EMT) of Plant Cells. Bio-protocol 7(19): e2564. DOI: 10.21769/BioProtoc.2564.
Category
Plant Science > Plant transformation > Ensifer-mediated transformation
Microbiology > Microbe-host interactions > Bacterium
Molecular Biology > DNA > Transformation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










