- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Detection of Protein S-nitrosothiols (SNOs) in Plant Samples on Diaminofluorescein (DAF) Gels
Published: Vol 7, Iss 18, Sep 20, 2017 DOI: 10.21769/BioProtoc.2559 Views: 8180
Reviewed by: Swetha ReddyWenrong HeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2870 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1989 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1772 Views
Abstract
In plant cells, the analysis of protein S-nitrosothiols (SNOs) under physiological and adverse stress conditions is essential to understand the mechanisms of Nitric oxide (NO)-based signaling. We adapted a previously reported protocol for detecting protein SNOs in animal systems (King et al., 2005) for plant samples. Briefly, proteins from plant samples are separated via non-reducing SDS-PAGE, then the NO bound by S-nitrosylated proteins is released using UV light and, finally, the NO is detected using the fluorescent probe DAF-FM (Rodriguez-Ruiz et al., 2017). Thus, the approach presented here provides a relatively quick and economical procedure that can be used to compare protein SNOs content in plant samples and provide insight in NO-based signaling in plants.
Keywords: Nitric oxideBackground
Nitric oxide (NO) is a free radical which can interact with a diverse array of biomolecules including proteins, lipids, and nucleic acids. In the case of proteins, one of the most relevant post-translational modifications (PTMs) is the covalent attachment of an NO group to the thiol (-SH) side chain of cysteine (Cys) present in peptides or proteins. This modification generates a family of NO-derived molecules called S-nitrosothiols (SNOs) which are important compounds in both animal and plant systems (Foster et al., 2003; Lindermayr and Durner, 2009; Astier et al., 2011; Broniowska and Hogg, 2012). Although this PTM is often designated as S-nitrosylation, the more appropriate term is S-nitrosation. It is difficult to detect, quantify and identify protein SNOs in plant systems. While there are several techniques to detect SNOs such as chemiluminescence, the biotin switch method, mass spectrometry, fluorescence detection, and antibody detection (against S-nitrosocysteine) (Kettenhofen et al., 2007; Foster, 2012; Devarie-Baez et al., 2013; Diers et al., 2014; Barroso et al., 2016; Mioto et al., 2017) many of these techniques require tedious sample preparation procedures that are time consuming and require sophisticated, expensive equipment.
Materials and Reagents
- 10-cm-diameter polystyrene Petri dishes (Fisher Scientific, catalog number: 12654785 )
- Parafilm M All-Purpose Paraffin Wax Film (Bemis, catalog number: PM996 )
- Sweet green pepper fruits were provided by Syngenta Seeds S.A. (El Ejido, Spain)
Note: This company grows pepper plants in experimental glass-covered greenhouses under optimal conditions of light, temperature and humidity.
- Arabidopsis thaliana ecotype Columbia seeds (originally obtained from NASC, Nottingham Arabidopsis Stock Center)
- Ethanol (Fisher Scientific, catalog number: 10517694 )
- Commercial Bleach (20%)
- Murashige and Skoog medium (Sigma-Aldrich, catalog number: M5524 )
- Sucrose (Sigma-Aldrich, catalog number: 84097 )
- Phyto-agar (Sigma-Aldrich, catalog number: P8169-100G )
- Bio-Rad Protein Assay Dye Reagent (Bio-Rad Laboratories, catalog number: 5000006 )
- Bovine serum albumin (BSA) Fraction V (Roche Diagnostics, Sigma-Aldrich, catalog number: 10735078001 )
- 4-20% Precast TGX Mini-Protean gel (Bio-Rad Laboratories, catalog number: 4561093 )
- Ascorbate (AsA) (Sigma-Aldrich, catalog number: A7631-25G )
- Copper(I) chloride (CuCl) (Sigma-Aldrich, catalog number: 651745-5G )
- N-ethylmaleimide (NEM) (Sigma-Aldrich, catalog number: E3876-5G )
- Dithiothreitol (DTT) (Roche Diagnostics, catalog number: 10708984001 )
- Reduced glutathione (GSH) (Sigma-Aldrich, catalog number: G4251-5G )
- β-Mercaptoethanol (ME) (Sigma-Aldrich, catalog number: M6250-10ML )
- Tris (AMRESCO, catalog number: 0497 )
- Ethylenediaminetetraacetic acid, disodium salt, dihydrate (Na2-EDTA) (Sigma-Aldrich, catalog number: E5134 )
- Triton X-100 (AMRESCO, catalog number: 0694 )
- Glycerol (AMRESCO, catalog number: E520 )
- Sodium dodecyl sulfate (SDS; electrophoresis grade)
- Bromophenol blue (Sigma-Aldrich, catalog number: B0126-25G )
- 3-Amino,4-aminomethyl-2’,7’-difluorescein (DAF-FM) (Sigma-Aldrich, catalog number: D2196 )
- Grinding buffer (see Recipes)
- Sample treatment buffer (2x) (see Recipes)
- Standard running buffer for SDS-PAGE containing 1 mM EDTA (see Recipes)
- Gel staining solution (see Recipes)
Equipment
- Set of Gilson micropipettes (Gilson, P10, P20 and P100)
- Plant growth cabinet (Panasonic Biomedical, model: MLR-352-PE )
- Porcelain mortar and pestle (VWR, catalog numbers: 410-0110 and 410-0120 , respectively)
- Refrigerated centrifuge Hettich Mikro 220R (Hettich Lab Technology, model: Mikro 220 R , catalog number: 2205)
- Vertical Slab gels Electrophoresis System (Bio-Rad Laboratories, model: Mini-PROTEAN®, catalog number: 1658003EDU )
- Standard UV-transilluminator (302-312 nm), used in molecular biology laboratory
- Molecular Imager PharosFX system (Bio-Rad Laboratories, model: PharosFXTM, catalog number: 1709460 )
Note: This product has been discontinued.
- EvolutionTM 201 UV-visible spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: EvolutionTM 201 , catalog number: 912A0890)
Software
- ImageJ (free application available in https://imagej.net/)
Procedure
- Preparation of plant extracts
- Sweet green pepper (Capsicum annuum) fruits were provided by Syngenta Seeds S.A. (El Ejido, Spain) from plants grown in experimental greenhouses with optimal nutrients supplementation applied on rockwood as substrate.
- Surface-sterilize Arabidopsis thaliana ecotype Columbia seeds for 5 min in 70% ethanol containing 0.1% SDS. Then, place in sterile water containing 20% (v/v) commercial Bleach and 0.1% SDS for 20 min. Wash four times with sterile water. Grow seeds on Petri plates over commercial Murashige and Skoog medium (Sigma-Aldrich) at a pH of 5.5, containing 1% (w/v) sucrose and 0.8% (w/v) phyto-agar. Place the seedlings for 14 days at 16 h light, 22 °C/8 h dark, 18 °C, under a light intensity of 100 μE m-2 sec-1.
- Homogenize plant samples (between 0.1 to 0.5 g) in a mortar and pestle with Gridding buffer (see Recipes) in a ratio 1:1 (w/v) for pepper fruits and ratio 1:3 (w/v) for Arabidopsis. Perform these operations at 0-4 °C, for example using a container with ice (see Figure 1).
Figure 1. Illustrative picture showing the Disposition of the mortar in the container with ice
- Centrifuge extracts at 27,000 x g at 4 °C for 20 min.
- Use the supernatants for the protein assays. Determine protein concentration with the Bio-Rad Protein Assay Dye Reagent (Bio-Rad Laboratories, Hercules, CA) using bovine serum albumin as the standard according with the ‘Microassay Procedure’ as outlined by the manufacturer.
- SDS-PAGE and S-nitrosothiols staining
- Pre-run SDS-PAGE using a precast 4-20% gradient TGX Mini-Protean gel (without samples) for 30 min at 30 mA per gel with standard running buffer containing 1 mM EDTA.
Note: The gel is pre-run primarily to remove any potential traces of unpolymerized acrylamide.
- Prepare samples with sample treatment buffer (see Recipes) in ratio 1:1 (v/v). For each samples, 25 µg of protein is loaded per lane.
- Run the electrophoresis at 15 mA per gel. The electrophoresis should be stopped when the front line indicated by the bromophenol blue (used for tracing the migration of samples) is 1 cm from the end of the gel (usually for 45 to 60 min).
- Wash the gel with 20-25 ml ultrapure water containing 1 mM EDTA for 5 min.
- Coat the gel with Gel staining solution (28 µM DAF-FM) for 10 min at room temperature in the dark.
Note: Cover with a piece of Parafilm of the same size and use a roller to gently smooth the staining solution over the gel such that there is a homogenous distribution of the solution.
- Expose the gel to UV light for 5 min.
- Take a picture with the fluor imager (PharosFMTM) according to the manufacturer’s instructions (excitation wavelength of 488 nm and emission wavelength of 530 nm) (see Figure 2).
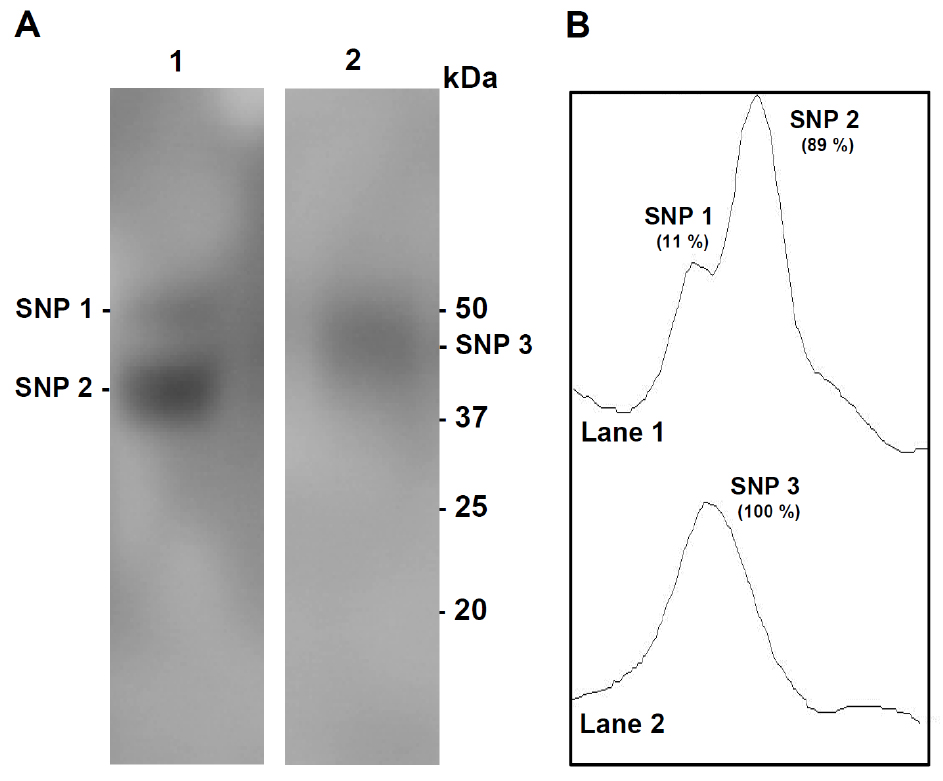
Figure 2. Detection and quantification of S-nitrosylated proteins in plant samples. A. Detection of endogenous S-nitrosylated (S-nitrosated) proteins (SNP) on DAF gels. Lane 1. Sweet green pepper fruits; Lane 2. Arabidopsis thaliana 14-day-old seedlings. Protein samples (25 µg) were separated by SDS-PAGE (gradient gels 4-20%) under non-reducing conditions. Molecular weight markers are indicated on the right. B. Densitometric scans of S-nitrosated proteins and its relative quantification (%) made by the ImageJ program
- Pre-run SDS-PAGE using a precast 4-20% gradient TGX Mini-Protean gel (without samples) for 30 min at 30 mA per gel with standard running buffer containing 1 mM EDTA.
Data analysis
Quantification of the S-nitrosated protein bands can be done by densitometric analysis, for example, using the ImageJ program (see Figure 2B).
Notes
Any techniques could have false positives. Therefore, it is highly recommended to perform several internal controls to avoid potential artifacts in new plant samples. Before loading samples on DAF gels it is recommended that plant samples be pre-treated at 25 °C for 3 h with different chemicals capable of: A) decomposing SNOs, such as 20 mM ascorbate (AsA) and 0.1 mM CuCl; B) blocking free thiols, such as 5 mM N-ethylmaleimide (NEM); and C) reducing agents, such as 20 mM dithiothreitol (DTT), 20 mM reduced glutathione (GSH) and 100 mM β-mercaptoethanol (ME).
Recipes
- Grinding buffer
50 mM Tris-HCl, pH 7.8
0.1 mM EDTA
0.2% (v/v) Triton X-100
10% (v/v) glycerol
- Sample treatment buffer (2x)
250 mM Tris-HCl, pH 6.8
8% (w/v) SDS
40% (w/v) glycerol
0.006% (w/v) bromophenol blue
- Standard running buffer for SDS-PAGE containing 1 mM EDTA
Running buffer: 0.375 mM Tris-HCl, pH 8.8
1 mM EDTA
- Gel staining solution
28 µM DAF-FM solution prepared in ultrapure water
Note: Prepare before use and protected from the light.
Acknowledgments
MRR acknowledges an FPI contract (BES-2012-055904) from the Ministry of Economy and Competitiveness, Spain. This work has been supported by the ERDF co-financed grant AGL2015-65104-P from the Ministry of Economy and Competitiveness, Spain. The authors are also grateful for the previous work done by King et al. (2005) which has been of great value to adapt it to plant samples.
References
- Astier, J., Rasul, S., Koen, E., Manzoor, H., Besson-Bard, A., Lamotte, O., Jeandroz, S., Durner, J., Lindermayr, C. and Wendehenne, D. (2011). S-nitrosylation: an emerging post-translational protein modification in plants. Plant Sci 181(5): 527-533.
- Barroso, J. B., Valderrama, R., Carreras, A., Chaki, M., Begara-Morales, J. C., Sanchez-Calvo, B. and Corpas, F. J. (2016). Quantification and localization of S-nitrosothiols (SNOs) in higher plants. Methods Mol Biol 1424: 139-147.
- Broniowska, K. A. and Hogg, N. (2012). The chemical biology of S-nitrosothiols. Antioxid Redox Signal 17(7): 969-980.
- Devarie-Baez, N. O., Zhang, D., Li, S., Whorton, A. R. and Xian, M. (2013). Direct methods for detection of protein S-nitrosylation. Methods 62:171-176.
- Diers, A. R., Keszler, A. and Hogg, N. (2014). Detection of S-nitrosothiols. Biochim Biophys Acta 1840(2): 892-900.
- Foster, M. W. (2012). Methodologies for the characterization, identification and quantification of S-nitrosylated proteins. Biochim Biophys Acta 1820(6): 675-683.
- Foster, M. W., McMahon, T. J. and Stamler, J. S. (2003). S-nitrosylation in health and disease. Trends Mol Med 9(4): 160-168.
- Kettenhofen, N. J., Broniowska, K. A., Keszler, A., Zhang, Y. and Hogg, N. (2007). Proteomic methods for analysis of S-nitrosation. J Chromatogr B Analyt Technol Biomed Life Sci 851(1-2): 152-159.
- King, M., Gildemeister, O., Gaston, B. and Mannick, J. B. (2005). Assessment of S-nitrosothiols on diaminofluorescein gels. Anal Biochem 346(1): 69-76.
- Lindermayr, C. and Durner, J. (2009). S-Nitrosylation in plants: pattern and function. J Proteomics 73(1): 1-9.
- Mioto, P. T., Rodriguez-Ruiz, M., Mot, A. C., Zuccarelli, R., Corpas, F. J., Freschi, L. and Mercier, H. (2017). Alternative fluorimetric-based method to detect and compare total S-nitrosothiols in plants. Nitric Oxide 68: 7-13.
- Rodriguez-Ruiz, M., Mioto, P., Palma, J. M. and Corpas, F. J. (2017). S-nitrosoglutathione reductase (GSNOR) activity is down-regulated during pepper (Capsicum annuum L.) fruit ripening. Nitric Oxide 68: 51-55.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Rodríguez-Ruiz, M., Mioto, P. T., Palma, J. M. and Corpas, F. J. (2017). Detection of Protein S-nitrosothiols (SNOs) in Plant Samples on Diaminofluorescein (DAF) Gels. Bio-protocol 7(18): e2559. DOI: 10.21769/BioProtoc.2559.
Category
Plant Science > Plant biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Electrophoresis
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











