- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Primary Culture System for Germ Cells from Caenorhabditis elegans Tumorous Germline Mutants
Published: Vol 7, Iss 15, Aug 5, 2017 DOI: 10.21769/BioProtoc.2424 Views: 9182
Reviewed by: Neelanjan BoseAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of Bacteria Residing in Caenorhabditis elegans Intestine
M. Fernanda Palominos and Andrea Calixto
May 5, 2020 5799 Views

Selective Enrichment and Identification of Cerebrospinal Fluid-Contacting Neurons In Vitro via PKD2L1 Promoter-Driven Lentiviral System
Wei Tan [...] Qing Li
Nov 20, 2025 1343 Views

Revisiting Primary Microglia Isolation Protocol: An Improved Method for Microglia Extraction
Jianwei Li [...] Guohui Lu
Dec 5, 2025 1475 Views
Abstract
The Caenorhabditis elegans germ line is an important model system for the study of germ stem cells. Wild-type C. elegans germ cells are syncytial and therefore cannot be isolated in in vitro cultures. In contrast, the germ cells from tumorous mutants can be fully cellularized and isolated intact from the mutant animals. Here we describe a detailed protocol for the isolation of germ cells from tumorous mutants that allows the germ cells to be maintained for extended periods in an in vitro primary culture. This protocol has been adapted from Chaudhari et al., 2016.
Keywords: C. elegansBackground
C. elegans hermaphrodite germ cells are generated in two adult stem cell niches located in the distal regions of the two gonad arms (Hansen and Schedl, 2013; Kimble and Seidel, 2013). In wild-type hermaphrodites, mitotic germ cells are restricted to the distal, stem cell niche regions of the gonad arms. Wild-type germ cells are syncytial, and contain an opening to a common cytoplasm that extends through the central region of the gonad arms. C. elegans tumorous germline mutants have increased mitotic proliferation of germ cells throughout the gonad. We discovered that tumorous germline mutants generally have fully cellularized germ cells that contain intact plasma membranes (Chaudhari et al., 2016). This cellularization allows the isolation of the germ cells and their maintenance in culture. This protocol describes the methodology and tissue culture medium to isolate and maintain germ cells from tumorous mutants in culture. While a culture medium has been described for the primary culture of C. elegans embryonic and larval cells (Strange et al., 2007; Zhang and Kuhn, 2013), germ cells do not survive in this medium (Chaudhari et al., 2016). We created a culture medium for germ cells that is called CeM1 for ‘C. elegans medium 1’. We anticipate that other iterations of the medium could be given subsequent numbers, e.g., ‘CeM2’. This protocol, first reported in Chaudhari et al., 2016, allows the isolation of essentially pure populations of germ cells and their maintenance in in vitro primary cultures. This culture system can allow new experimental approaches to probe germ cell biology in C. elegans.
Materials and Reagents
- Polystyrene tubes, 15 ml (Corning, Falcon®, catalog number: 352099 )
- Polypropylene tubes, 15 ml (Corning, catalog number: 25319-15 )
- Polypropylene tubes, 50 ml (Corning, catalog number: 25330-50 )
- Filter pipette tips, 100-1,000 µl (Corning, catalog number: 4809 )
- Filter pipette tips, 20-200 µl (Fisher Scientific, catalog number: 02-707-430 )
- Filter pipette tips, 2-20 µl (Fisher Scientific, catalog number: 02-707-435 )
- Filter pipette tips, 0.1-10 µl (Fisher Scientific, catalog number: 02-707-439 )
- Filter units 150 ml, PES 0.22 µm (EMD Millipore, catalog number: SCGPU01RE )
- Filter units 500 ml, PES 0.22 µm (EMD Millipore, catalog number: SCGPU05RE )
- Aluminum foil
- Tissue culture dish, 35 x 10 mm (Corning, Falcon®, catalog number: 353001 )
- Tissue culture dish, 24-well (Corning, catalog number: 3524 )
- Tissue culture dish, 96-well (Corning, catalog number: 3596 )
- Cell scrapers, 39 cm, disposable (SARSTEDT, catalog number: 83.1831 )
- Serological pipets, disposable 10 ml (Fisher Scientific, catalog number: 13-678-11E )
- Tissue culture flask, 12.5 cm2 (Corning, Falcon®, catalog number: 353018 )
- Platinum wire, 0.25 mm (Alfa Aesar, catalog number: 10288 )
- Needles, 21 G x 1 ½ (BD, catalog number: 305167 )
- Parafilm
- 0.5 ml centrifuge tube
- Microcentrifuge tubes, 1.5 ml (Fisher Scientific, catalog number: 05-408-129 )
- Razor blades, single edge
- Paper towel
- C. elegans tumorous germline mutant strain ET507, daf-16(mu86) I; cki-2(ok2105) II; glp-1(ar202) III
- Escherichia coli strain OP50
Notes:- We did not perform tests to determine how materials and reagents from other manufacturers function in germ cell isolation and culture, with the exception of the use of STARSTEDT 96-well plates (non-tissue culture-treated) (STARSTEDT, catalog number: 82.1581.001 ) for the in vitro culture of germ cells, which resulted in the premature death of the germ cells.
- C. elegans germline tumor mutant strain ET507 (as well as other tumorous mutant strains) and E. coli bacteria OP50 are available from the Caenorhabditis Genetics Center (CGC), http://www.cgc.cbs.umn.edu.
- We did not perform tests to determine how materials and reagents from other manufacturers function in germ cell isolation and culture, with the exception of the use of STARSTEDT 96-well plates (non-tissue culture-treated) (STARSTEDT, catalog number: 82.1581.001 ) for the in vitro culture of germ cells, which resulted in the premature death of the germ cells.
- Fetal bovine serum (FBS) (Atlanta Biologicals, catalog number: S11550 )
- Amberlite IRA 400-CL (Sigma-Aldrich, catalog number: 247669 )
- Charcoal-dextran (Sigma-Aldrich, catalog number: C6241 )
- Liquid nitrogen
- Phosphate buffered saline (PBS) (GE Healthcare, HyCloneTM, catalog number: SH30256.01 )
- Schneider’s insect medium (Thermo Fisher Scientific, GibcoTM, catalog number: 21720024 )
- Leibovitz’s L-15 medium without phenol red (Thermo Fisher Scientific, GibcoTM, catalog number: 21083027 )
- Penicillin/streptomycin (Sigma-Aldrich, catalog number: P4333 )
- Hemin chloride (MP Biomedicals, catalog number: 0219402501 )
- RPMI Vitamins (Sigma-Aldrich, catalog number: R7256 )
- L-glutathione, reduced (Sigma-Aldrich, catalog number: G4251 )
- Normocin (InvivoGen, catalog number: ant-nr-1 )
- Trehalose (Sigma-Aldrich, catalog number: T0167 )
- Osmolality standard, 100 mmol/kg (Wescor, catalog number: OA-010 )
- Osmolality standard, 290 mmol/kg (Wescor, catalog number: OA-029 )
- Osmolality standard, 1,000 mmol/kg (Wescor, catalog number: OA-100 )
- Water, molecular biology grade (GE Healthcare, HyCloneTM, catalog number: SH30538.02 )
- Tryptone (Fisher Scientific, catalog number: BP1421-500 )
- Yeast extract (Fisher Scientific, catalog number: BP9727-2 )
- Sodium chloride (NaCl) (Avantor Performance Materials, J.T. Baker®, catalog number: 3624-05 )
- Sodium hydroxide (NaOH) (Avantor Performance Materials, J.T. Baker®, catalog number: 3728-01 )
- Bacto-peptone (BD, BactoTM, catalog number: 211677 )
- Agar (RPI, catalog number: A20020-5000 )
- Cholesterol (Avantor Performance Materials, J.T. Baker®, catalog number: 1580-01 )
- Ethanol, 100% (used to make 70% with distilled water) (Decon Labs, catalog number: 2716 )
- Magnesium sulfate, anhydrous (MgSO4) (Avantor Performance Materials, J.T. Baker®, catalog number: 2506-01 )
- Calcium chloride (CaCl2) (Avantor Performance Materials, J.T. Baker®, catalog number: 1313-01 )
- Potassium phosphate, monobasic (KH2PO4) (Avantor Performance Materials, J.T. Baker®, catalog number: 3246-05 )
- Sodium phosphate, heptahydrate (Na2HPO4·7H2O) (Fisher Scientific, catalog number: S373-3 )
- Sodium hypochlorite, 6% (RICCA Chemical, catalog number: 7495.7-32 )
- Tetracycline (Sigma-Aldrich, catalog number: 87128 )
- Chloramphenicol (RPI, catalog number: C61000-25.0 )
- Kanamycin (Fisher Scientific, catalog number: BP906-5 )
- Ethidium homodimer (Biotium, catalog number: 40010 )
- Dimethyl sulfoxide (DMSO) (Fisher Scientific, catalog number: D128-500 )
- Hoechst 33342 (Sigma-Aldrich, catalog number: B2261 )
- Calcein-AM (Biotium, catalog number: 80011 )
- 2xYT bacterial medium (see Recipes)
- LB bacterial medium (see Recipes)
- 3x NGM agar plates seeded with OP50 bacteria (see Recipes)
- M9 buffer (see Recipes)
- Sodium hypochlorite solution (see Recipes)
- Platinum-wire worm pick (see Recipes)
- Tetracycline stock (see Recipes)
- Chloramphenicol stock (see Recipes)
- Kanamycin stock (see Recipes)
- Cholesterol stock (see Recipes)
- Antibiotic-enriched PBS with heat-killed bacteria (see Recipes)
- Hemin chloride stock (see Recipes)
- Stock of Hoechst 33342, calcein-AM, and ethidium homodimer (see Recipes)
Equipment
- Temperature-controlled water bath
- Table-top centrifuge with swinging bucket rotor for 15 ml and 50 ml tubes
- Rotator, single speed (Barnstead Thermolyne, catalog number: C415110 )
- 500 ml plastic bottle
- Freezing point osmometer (Advanced Digimatic Osmometer 3DII, Advanced Instruments, model: Model 3D2 )
- Low-temperature incubator
- Bottom illuminated stereomicroscope with frosted glass stage or frosted mirror (various Nikon or Leica models)
- Airtight containers (LockandLock, various size Lock & Lock containers available from Amazon)
- Phase-contrast inverted compound microscope (various Nikon, Leica, or Olympus models with 10x and 20x objectives)
- Nutator (BD, catalog number: 421105 )
- Hemacytometer (Reichert Bright-Line, Hauser Scientific, catalog number: 1492 )
- Upright fluorescence compound microscope (various Nikon, Leica, or Olympus models with 5x, 20x, 40x, and 64x objectives)
- 2 L Erlenmeyer flasks (Corning, PYREX®, catalog number: 4980-2L )
- Autoclave
- 50 ml Erlenmeyer flasks (Corning, PYREX®, catalog number: 4980-50 )
- Plastic 1 L measuring cup with handle (Fisher Scientific, catalog number: 02-543-36C )
- Stir bar
- Bottles, 125 ml, glass (WHEATON, catalog number: 219815 )
- Bunsen burner
- Analytical balance
- pH meter
- Pipette controller (Pipet-Aid) (Drummond Scientific, catalog number: 4-000-110 )
- Pasteur pipets, 9 inch, glass (Fisher Scientific, catalog number: 22-063172 )
- Bulbs for Pasteur pipets (Fisher Scientific, catalog number: 03-448-21 )
- Laminar-flow tissue culture hood
- Aspirator
Procedure
- Pretreatment of FBS
- If the FBS is stored frozen in aliquots (at -20 °C or -80 °C), thaw the appropriate amount of FBS to make the desired volume of CeM1 medium (FBS is used at a final concentration of 8%). Note, pretreatment of FBS leads to a loss of ~20% of the volume due to trapping of FBS in charcoal-dextran, e.g., 25 ml of FBS will produce ~20 ml of pre-treated FBS. FBS can be thawed at room temperature or in a 37 °C water bath. The pretreatment contains two overnight incubation steps. The incubation of FBS with Amberlite IRA 400-CL beads and charcoal-dextran removes hydrophobic molecules, such as steroid hormones. The pretreatment steps are required to maintain germ cell viability in the CeM1 medium (Chaudhari et al., 2016). It is not known how the pretreatments alter the FBS to improve cell survival.
- Heat inactivate the thawed FBS by incubation at 56-65 °C for 30 min in a temperature-controlled water bath with occasional swirling (every 5-10 min).
- Prewash an amount of Amberlite IRA 400-CL beads that is based on a final concentration of 50 mg of beads per ml of FBS that will be subject to pretreatment; for 25 ml of FBS, this would be 1.25 g of beads. For the prewash, place the beads in a 15 ml or 50 ml polypropylene tube. The beads are prewashed by adding molecular-grade water to the tube (~13 or ~45 ml depending on whether a 15 ml or 50 ml tube is used) and inverting the tube several times. The beads are collected by centrifugation at 930 x g for 1 min in a table-top centrifuge. Aspirate the water above the beads. Repeat the wash two times. Remove residual water between the beads by placing a 1,000 µl pipet tip against the bottom of the tube and pipetting out the water.
- Add the heat-inactivated FBS to the tube with the washed Amberlite IRA 400-CL beads.
- Rotate the FBS and Amberlite IRA 400-CL beads for 4-6 h at room temperature in a single-speed rotator.
- Pre-wash a second set of Amberlite IRA 400-CL beads (1.25 g for 25 ml of FBS), as was done in step A3.
- Spin down the beads (930 x g for 1 min), and transfer the FBS to a second batch of washed Amberlite IRA 400-CL beads in a second tube.
- Rotate the FBS and beads overnight at 4 °C in the rotator.
- The next day, prepare charcoal-dextran for incubation with the FBS. The amount of charcoal-dextran is based on a final concentration of 100 mg of charcoal-dextran per ml of FBS; for 25 ml of FBS, this would be 2.5 g of charcoal-dextran. Precrush the charcoal-dextran against the side of the weigh dish–to break-up clumps. Place in a 15 ml or 50 ml polypropylene tube.
- Spin down the Amberlite IRA 400-CL beads (930 x g for 1 min), and transfer the FBS to the tube with the charcoal-dextran.
- Rotate the FBS with the charcoal-dextran overnight at 4 °C in the rotator.
- Remove the charcoal-dextran with two sequential centrifugations, each at 2,850 x g for 30 min, 4 °C, followed by transferring FBS to a new polypropylene tube (leaving a small fraction, ~100 µl, over the pellet to ensure that no residue from the pellet is taken).
- FBS that has been treated by heat-inactivation, and incubations with Amberlite IRA 400-CL and charcoal-dextran can either be used immediately, stored at 4 °C for one to two weeks, or stored for longer periods (months or a few years) at -80 °C (after snap-freezing aliquots in liquid nitrogen).
Notes:
- Different lots of FBS have large effects on the long-term survival of germ cells in culture. It would be advantageous to obtain samples of multiple lots from FBS manufacturers/distributors to test for germ cell survival prior to purchasing a specific lot of FBS.
- Throughout the protocol, the centrifuge speeds are based on a Beckman Coulter Allegra X-15R table-top centrifuge with a swinging bucket rotor, which has the following rpm to rcf (x g) conversions: 300 rpm = 21 x g; 1,000 rpm = 230 x g; 2,000 rpm = 930 x g; 3,500 rpm = 2,850 x g; 4,000 rpm = 3,700 x g.
- If the FBS is stored frozen in aliquots (at -20 °C or -80 °C), thaw the appropriate amount of FBS to make the desired volume of CeM1 medium (FBS is used at a final concentration of 8%). Note, pretreatment of FBS leads to a loss of ~20% of the volume due to trapping of FBS in charcoal-dextran, e.g., 25 ml of FBS will produce ~20 ml of pre-treated FBS. FBS can be thawed at room temperature or in a 37 °C water bath. The pretreatment contains two overnight incubation steps. The incubation of FBS with Amberlite IRA 400-CL beads and charcoal-dextran removes hydrophobic molecules, such as steroid hormones. The pretreatment steps are required to maintain germ cell viability in the CeM1 medium (Chaudhari et al., 2016). It is not known how the pretreatments alter the FBS to improve cell survival.
- Preparation of CeM1 medium
- Prepare CeM1 medium in a bottle that has not been washed with soap (e.g., a 500 ml plastic bottle that previously held water, PBS, or tissue culture medium, and that was cleaned by rinsing with distilled water and air drying). If less than 50 ml of medium is prepared, it can be made in a 50 ml polypropylene tube.
- The percentages/concentrations of CeM1 ingredients are described in Table 1. Initially, add Schneider’s insect medium, Leibovitz’s L-15 medium without phenol red, pretreated FBS, penicillin/streptomycin, RPMI vitamins, and normocin.
Table 1. CeM1 medium components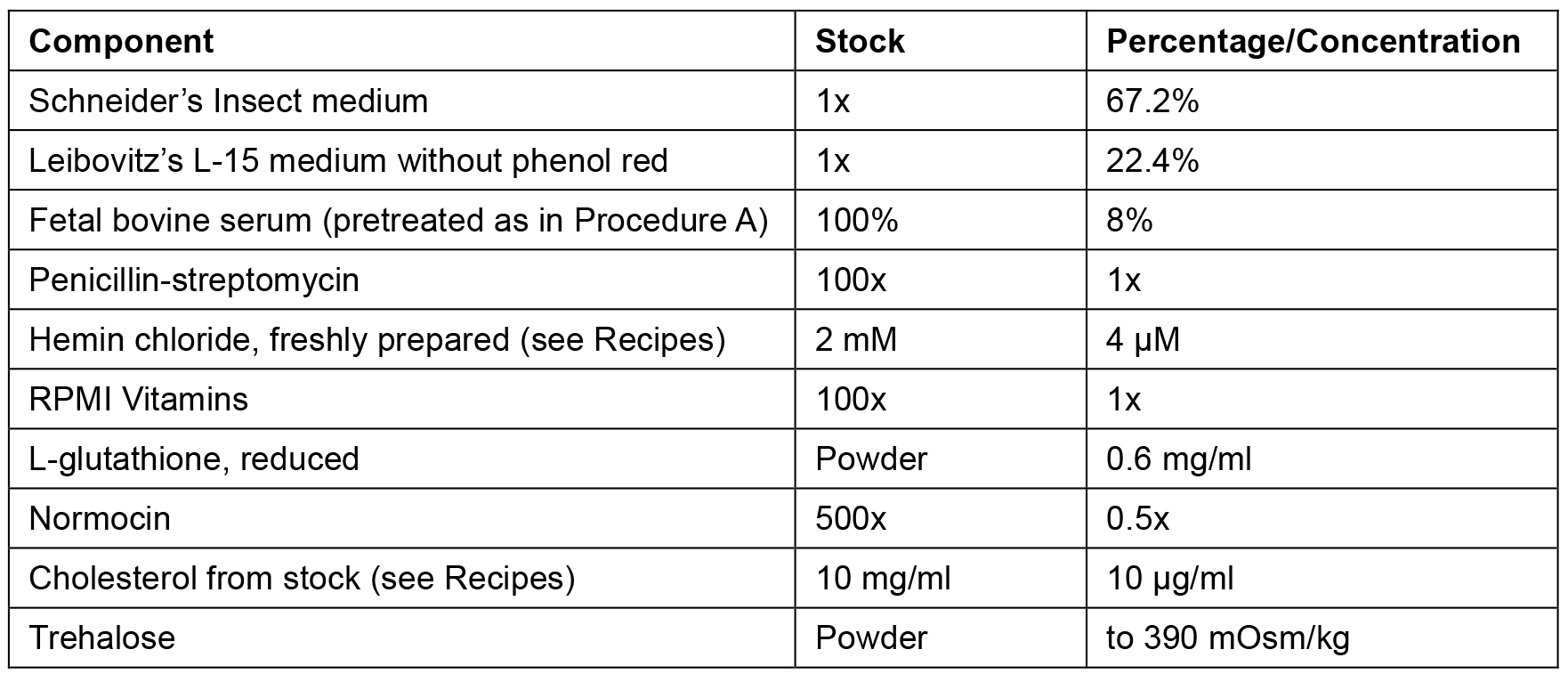
- Add cholesterol from a 10 mg/ml stock in ethanol (see Recipes).
- Add trehalose at 18 mg/ml, and swirl the bottle to dissolve the powder. This should produce an osmolality that is lower than the desired 390 mOsm.
- Add reduced L-glutathione as a powder and swirl the container to dissolve it.
- Add hemin chloride from a freshly-prepared 4 mM stock (see Recipes).
- Adjust the pH to 6.5 by adding 5 N NaOH.
- Prepare the freezing point osmometer by turning it on for 30 min prior to measurements and then calibrate it with 100, 290, and 1,000 mOsm standards.
- Determine the osmolality of the CeM1 medium with the freezing point osmometer. The osmolality should be below the desired osmolality of 390 mOsm.
- Adjust the osmolality of the CeM1 medium to 390 mOsm/kg by adding additional trehalose (dissolved by swirling).
- Sterile filter the CeM1 medium through 0.22 µm 150 or 500 ml filter units.
- Transfer the sterile CeM1 medium to 15 ml or 50 ml polypropylene tubes so that it can be stored in aliquots with little air in the container (to limit evaporation and condensation). Store the aliquot tubes at 4 °C, wrapped in aluminum foil to protect them from light. The aliquots can be used for at least two months.
- Prepare CeM1 medium in a bottle that has not been washed with soap (e.g., a 500 ml plastic bottle that previously held water, PBS, or tissue culture medium, and that was cleaned by rinsing with distilled water and air drying). If less than 50 ml of medium is prepared, it can be made in a 50 ml polypropylene tube.
- Preparation of heat-killed OP50 bacteria
- Grow one liter of OP50 bacteria in 2x YT bacterial medium overnight at 37 °C.
- Spin down the bacteria at 3,700 x g for 30 min at 4 °C, decant the liquid.
- Resuspend in 5 ml of 0.9% NaCl and place in a 15 ml polypropylene tube.
- Incubate the tube in a water bath set at 62-67 °C for 24 h.
- Store heat-killed bacteria at 4 °C.
- Grow one liter of OP50 bacteria in 2x YT bacterial medium overnight at 37 °C.
- Preparation of tumorous mutants
- A source of eggs for the procedure can be a mixed-stage population of tumorous germline mutants that contains gravid adults (i.e., adults with eggs in their body) and laid eggs. One 10 cm 3x NGM agar plate seeded with OP50 bacteria (see Recipes) and a somewhat dense population of mixed-stage animals will generally be adequate to provide sufficient eggs for the procedure. If using a temperature sensitive (ts) tumorous germline mutant, the 3x NGM agar plate should be incubated at the permissive temperature of 15-16 °C in a low-temperature incubator to allow the mutants to produce eggs. Several hundred eggs is sufficient for most small-scale experiments, e.g., 25 adults (derived from eggs) is used for isolation of germ cells for every well of a 24-well plate (each with 0.5 ml of medium).
- Collect C. elegans animals and eggs from the plate by rinsing with M9 buffer and scraping the eggs in M9 buffer with a cell scraper. The scraping is carried out by tilting the plate with one hand at an approximate angle of 30°, which makes the M9 buffer settle to the bottom part of the tilted plate. The cell scraper then is used with the other hand to move the cells and bacteria into the M9 liquid. The plate is then rotated so that regions with intact bacteria are brought to the top of the plate where they can be scraped into the liquid. When all the bacteria and eggs are in the M9 buffer, the liquid is transferred with a sterile glass Pasteur pipet to a 15 ml polystyrene tube.
- Spin down the animals and eggs by centrifugation at 930 x g for 1 min.
- Aspirate the liquid, and add 8 ml of sodium hypochlorite solution (see Recipes) and incubate with frequent inversions for approximately 3.5-4.5 min. During the incubation, occasionally place the 15 ml tube on a stereomicroscope to observe the animals. The incubation with sodium hypochlorite should be continued until almost all of the adults have broken in half, which releases their eggs.
- Spin down the eggs at 930 x g for 1 min, aspirate solution.
- Wash four times with sterile M9 solution (each time filling up the 15 ml tube).
- Use a sterile Pasteur pipet to place the sterile eggs (in a small volume of M9) onto an OP50-seeded 3x NGM plate. Although not required, if a more synchronized population of adults is desired, the sterile eggs can instead be placed in 15 ml of M9 with 5 µg/ml cholesterol in a 10 cm Petri dish, and left overnight at room temperature so that all of the eggs hatch and the resulting larvae arrest at the L1 larval stage. The L1 larvae can then be collected in a polystyrene 15 ml tube using a glass Pasteur pipet, spun down, excess liquid aspirated, and the eggs in a small volume of M9 (~100 µl) added to an OP50-seeded 3x NGM plate.
- Place the 3x NGM plate with eggs at 25 °C in a low-temperature incubator for four days. During this time period, the animals will develop to become adults with germline tumors (Figure 1).
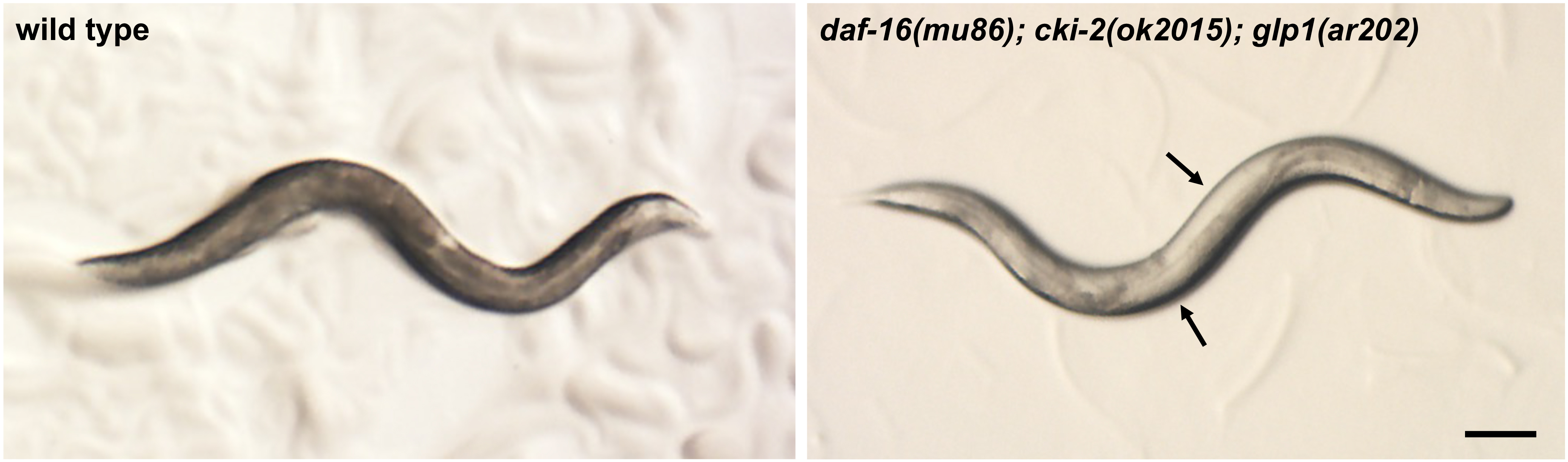
Figure 1. Stereomicroscope image of wild-type and tumorous mutant C. elegans adult hermaphrodites grown at 25 °C. Note that in the daf-16(mu86); cki-2(ok2105); glp-1(ar202) mutant, germline tumors are visible as white areas in the body (arrows). Scale bar = 100 µm. - Collect the germline tumorous mutant animals from the plate by adding M9 solution to the edge of the plate and transfer the floating animals off the plate with a sterile Pasteur pipet into a 15 ml polystyrene tube (without disturbing the bacterial lawn).
- Wash the collected mutant animals four times with ~14 ml of M9 buffer to remove live bacteria (spinning at 230 x g for 1 min for each wash).
- Add the washed animals to a 12.5 cm2 cell culture flask that contains 2.5 ml of antibiotic-enriched PBS with heat-killed bacteria (see Recipes).
- Close the flask tightly and incubate at 25 °C overnight in a low-temperature incubator.
Notes:
- If eggs fail to spin down in the sodium hypochlorite solution, then the density of the Bleach/sodium hypochlorite used in preparing the sodium hypochlorite solution is too high. In this case, after the initial incubation that dissolves the animals, add sterile water to fill the 15 ml tube and invert to mix prior to spinning down the eggs. This reduces the density of the solution so that the eggs will pellet.
- Occasionally, animals that will be used for germ cell isolation (prior to harvesting into antibiotic-containing PBS) are contaminated with fungus or bacteria other than OP50 (other bacteria generally appear either as denser bacterial colonies on the relatively thin OP50 bacterial lawn and/or as bacterial growths with altered color or texture). In this situation, we recommend aborting the experiment if long-term cultures are required, as continuing will generally lead to bacterial or fungal contamination in the in vitro culture. The contamination will generally not be overtly visible until the next day, and so isolated germ cells could be used for experiments within several hours of isolation.
- Polystyrene tubes are used to collect C. elegans larvae, adults, and eggs, because the animals do not stick to polystyrene. In contrast, if polypropylene tubes are used, animals and eggs will be lost during washes due to their sticking to the sides of the tube.
- Glass Pasteur pipets are used for transfers because eggs and animals do not stick to glass. In contrast, eggs and animals will stick to plastic pipet tips, so these should not be used.
- SARSTEDT cell scrapers are packaged as sterile and disposable, but they do not need to be sterile if they are used for collecting eggs prior to sodium hypochlorite treatment. In this situation, they can be reused after washing with soap.
- When scraping eggs from the plates, care should be taken to avoid scraping the edge of the plate. The sharp edge of the scraper hitting the elevated agar at the edge of the plate can chip off pieces of agar that would then be included with the eggs.
- The tumorous mutants are grown at 25 °C to activate the glp-1(ar202) ts allele. If tumorous germline mutant alleles are used that are not ts, then the animals can be grown at lower temperatures (e.g., 20 °C).
- A source of eggs for the procedure can be a mixed-stage population of tumorous germline mutants that contains gravid adults (i.e., adults with eggs in their body) and laid eggs. One 10 cm 3x NGM agar plate seeded with OP50 bacteria (see Recipes) and a somewhat dense population of mixed-stage animals will generally be adequate to provide sufficient eggs for the procedure. If using a temperature sensitive (ts) tumorous germline mutant, the 3x NGM agar plate should be incubated at the permissive temperature of 15-16 °C in a low-temperature incubator to allow the mutants to produce eggs. Several hundred eggs is sufficient for most small-scale experiments, e.g., 25 adults (derived from eggs) is used for isolation of germ cells for every well of a 24-well plate (each with 0.5 ml of medium).
- Germ cell isolation
- Collect the animals from the antibiotic incubation into a 15 ml polystyrene tube and spin down (all spins with adult animals are 230 x g for 1 min).
- Wash the animals four times with sterile PBS (filling the tube).
- Wash one time with 5 ml of CeM1 medium.
- In a tissue culture hood, resuspend the washed animals in 2 ml of CeM1 medium and place in a 35 mm tissue culture dish.
- Prepare a second 35 mm tissue culture dish with a 120 µl circle of CeM1 medium in the center of the dish, which will be used for the isolation of germ cells.
- Take the plates out of the tissue culture hood to a bottom-illuminated stereomicroscope. While observing the animals through the stereomicroscope, transfer live animals (which move on their own or when prodded) using a platinum-wire worm pick from the 2 ml plate to the 120 µl circle of CeM1 medium in the second plate. Move the desired number of animals to the 120 µl circle (generally, 25 adult hermaphrodites are used per well of a 24-well plate, or 8 per well of a 96-well plate; we have harvested germ cells from up to 240 animals per one 120 µl circle). The worm pick for the transfer is a straightened platinum-wire with a flattened end, with a slight bend prior to the end so that when the wire is placed below multiple worms and lifted straight up, the worms are draped over both sides of the wire as it lifts out of the medium. The worms on the pick are then transferred to the media circle by placing the worms in the media and then dislodging them with a slight back and forth motion. Any non-moving worms that were transferred are removed with the flattened end of the wire pick.
- While visualizing through the stereomicroscope, cut the animals in the 120 µl circle into quarters using 21 gauge needles. To cut animals, one needle is placed on one side of an animal and the other needle is placed on the other side of the animal and then slid through the animal (next to the first needle), producing a scissor-like cutting action (Figure 2). Intact gonads that become extruded from animals are also cut.
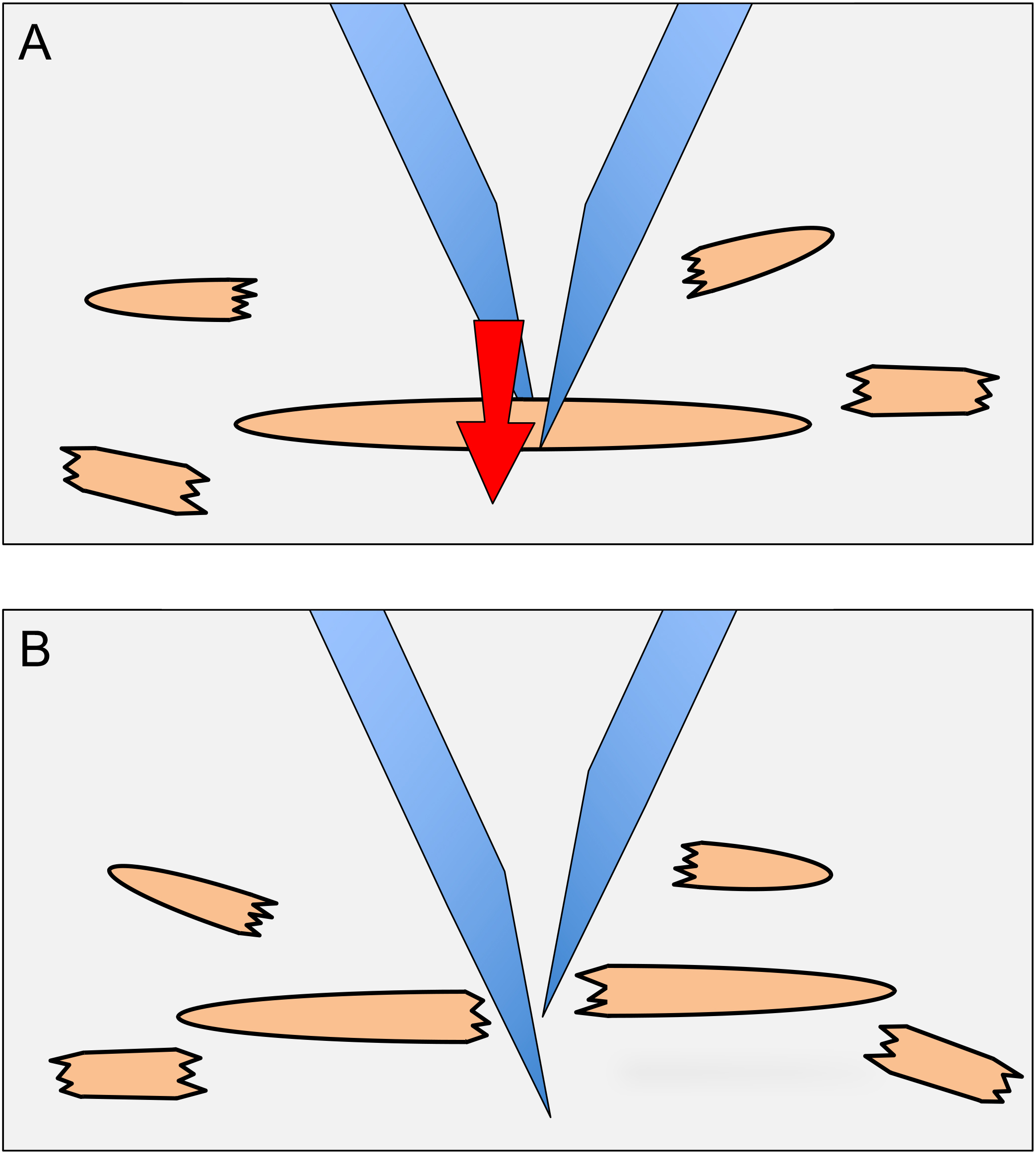
Figure 2. Dissection of animals to release germ cells. To dissect animals, needles are placed on either side of an animal as shown in (A). The needle in the background is brought forward next to the forward needle (arrow direction in A). The scissor-like action results in the dissection of the animal (B). The halves of the animal are then each dissected once more to produce smaller body pieces (as shown around the main animal). - Bring the plate back to the tissue culture hood. Add 1 ml of CeM1 medium to the plate with a 1,000 µl pipet tip, while holding the plate at ~30° angle. Pick up the 1 ml of CeM1 with the pipet and eject it forcefully into the middle of the plate (where the plastic has been scored/scratched by the needles); repeat this a total of 20 times. Do not release all the medium from the pipet when ejecting, as this will generate bubbles that can kill cells due to the surface tension on the bubbles.
- Transfer all the liquid from the plate to the bottom of a 15 ml polypropylene tube.
- Pipet a second 1 ml of CeM1 medium eight times against the center of the plate (still held at ~30° angle), and add it to the same 15 ml tube.
- Add a third 1 ml of CeM1 medium directly to the 15 ml tube, and pipet the liquid up and down ~10 times (again avoiding pipetting all the liquid out of the pipet tip to prevent the formation of bubbles).
- Spin the tube at 21-230 x g for 1 min to pellet body parts and large cell aggregates. The choice of centrifuge speed affects the distribution of germ cells isolated: spinning at 21 x g will retain large cell aggregates in the supernatant; spinning at 230 x g will remove large cell aggregates, leaving only individual cells and small cell aggregates in the supernatant.
- Transfer the supernatant to a new 15 ml polypropylene tube and spin for 930 x g for 5 min to pellet the cells.
- Aspirate the supernatant, and resuspend the cell pellet in fresh CeM1 medium by gentling pipeting up and down with a 1,000 µl pipet tip that has a relatively wide opening (as found on the specified Corning pipet tips). The cells are transferred to tissue culture dish(es) based on the number of animals harvested. Typically, 0.08 ml is used for each well of a 96-well plate (with 8 animals/well); 0.5 ml for each well of a 24-well plate (with 25 animals/well), and 2 ml for culturing in a 35 mm tissue culture dish (with > 50 animals/dish). The ratio of cells to medium can be changed based on the experiment.
- If multi-well tissue culture dishes are used and the outside wells do not contain cells, then fill the outside wells with sterile PBS to limit evaporation from wells that contain isolated germ cells.
- Seal the edge of the tissue culture dish or plate with Parafilm (to limit evaporation).
- Incubate the plates at 25 °C in a low-temperature incubator in an airtight container.
- Viable germ cells can be maintained for periods of up to four weeks.
- Visualize isolated germ cells with a phase-contrast inverted microscope (Figure 3).
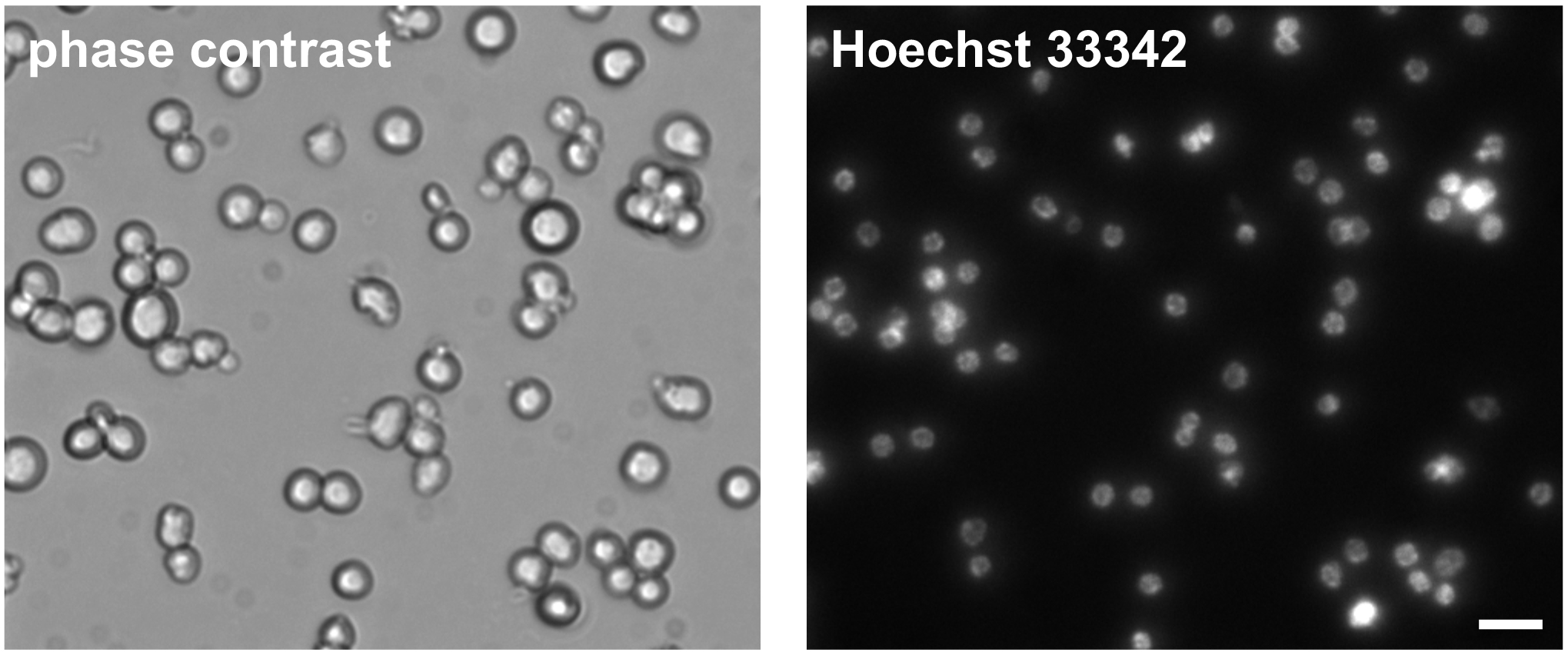
Figure 3. Phase-contrast and Hoechst 33342 DNA stain of isolated C. elegans germ cells. Cells were imaged one day post isolation. Scale bar = 10 µm.
- Collect the animals from the antibiotic incubation into a 15 ml polystyrene tube and spin down (all spins with adult animals are 230 x g for 1 min).
Data analysis
The numbers of live germ cells that are isolated in vitro can be determined as described in Chaudhari et al., 2016. To count live cells, the cells are stained with the live-cell stain calcein-AM, the dead-cell stain ethidium homodimer, and the DNA stain Hoechst 33342. Calcein-AM can freely diffuse into live cells but is then converted by cellular esterases to a fluorescent form that is unable to cross intact plasma membranes and so remains within the cell. Ethidium homodimer stains dead cells that lack an intact plasma membrane. The cell permeable dye Hoechst 33342 allows the visualization of nuclear DNA. The procedure to count live and dead cells is as follows.
- Isolated germ cells are gently mixed in the tissue culture medium with a filtered 1,000 µl pipet tip.
- 40 µl of the cells is transferred to a 0.5 ml centrifuge tube with a new 1,000 µl pipet tip.
Note: A 1,000 µl pipet tip is recommended because the opening of the tip is large enough to prevent the cells from being damaged while they are drawn into the tip. - The stock mixture of Hoechst 33342, calcein-AM, and ethidium homodimer (see Recipes) is diluted 1:10 in CeM1, and then 1 µl of the dilution is mixed with the 40 µl of cells. The final concentration is 5 µg/ml Hoechst 33342; 1 µM calcein-AM; and 0.1 µM ethidium homodimer.
- The tube with the cells is wrapped in aluminum foil (to block light) and incubated on a nutator for 30 min at room temperature.
- A pipet tip for a 20 µl pipetter is sterilely cut ~5 mm from the tip with a razor blade (sterilized by washing with 70% ethanol) in order to make a larger opening, and is used to resuspend the stained cells, and then ~10 µl of the solution is loaded into a hemacytometer.
- The hemacytometer is analyzed on an upright compound fluorescence microscope. Live cells stain with calcein-AM (green) and contain nuclei that stain with Hoechst 33342 (blue), but lack staining with ethidium homodimer (red). Dead cells stain with ethidium homodimer, but lack staining with calcein-AM.
If hermaphrodites with large tumors are selected for dissection, up to 6,000 live germ cells can be isolated per hermaphrodite if a low speed is used for the initial spin (to remove animal parts and larger cell aggregates) (Chaudhari et al., 2016). The number of isolated germ cells will be reduced if higher speeds are used for the initial spin, as this will remove more aggregated germ cells. Analysis of the cells isolated from wild-type animals (from which live germ cells cannot be isolated due to their syncytial nature) vs. ET507 tumorous germline mutants showed that over 99% of the isolated cells from ET507 are germ cells (Chaudhari et al., 2016).
Recipes
- 2xYT bacterial medium
16 g tryptone
10 g yeast extract
5 g NaCl
1 L distilled water
Add ingredients to a 2 L Erlenmeyer flask, cover with aluminum foil, and autoclave for 30 min - LB bacterial medium
10 g tryptone
5 g yeast extract
10 g NaCl
0.2 ml 5 N NaOH
1,000 ml distilled water
Dissolve components by mixing with a stir bar. Aliquot to ten 100 ml bottles and autoclave for 20 min
Note: This recipe is the conventional recipe for LB bacterial medium, but it differs from the original LB medium (Bertani, 1951) in that it does not contain 0.1% glucose. - 3x NGM agar plates seeded with OP50 bacteria
- 3 g NaCl
7.5 g Bacto-peptone
20 g agar
1 ml 5 mg/ml cholesterol (dissolved in 100% ethanol)
1 ml 1 M MgSO4
1 ml 0.1 M CaCl2
972 ml distilled water
Add to a 2 L Erlenmeyer flask that is then covered with aluminum foil - Separately prepare a 50 ml Erlenmeyer flask or other small glass container (covered with aluminum foil) and filled with: 25 ml 1 M KH2PO4
- Autoclave the 2 L flask, small container, and a 1 L plastic pouring measuring cup with handle (covered with aluminum foil) for 30 min
- After autoclaving, add the 25 ml 1 M KH2PO4 to the 1 L in the flask. Swirl, pour into the 1 L measuring cup. Wait for the bubbles to disappear. Pour into 10 cm Petri dishes. Allow the agar plates to air dry for two days
- Grow an overnight culture of OP50 bacteria in LB medium. Put a small amount of the overnight OP50/LB culture onto each plate with a 10 ml glass pipet (that has a completely smooth tip), spread the bacteria on the surface of the plate with the glass pipet, leaving an ‘unseeded’ space around the edge of the plate
- Incubate the plates at room temperature for two days and then place in the plastic sleeves that originally held the Petri dishes, seal with tape, and store at 4 °C until needed. These plates can be used for one to two months
Note: This recipe is a modification of the conventional NGM recipe (Sulston and Hodgkin, 1988); it differs in that it contains a three-fold higher concentration of peptone to allow thicker growth of bacteria.
- M9 buffer
11.32 g Na2HPO4·7H2O
3 g KH2PO4
5 g NaCl
0.012 g MgSO4
Add distilled H2O to 1 L
Aliquot to ten 100 ml bottles and autoclave for 20 min
Note: This recipe is a modification of the conventional M9 buffer recipe (Sulston and Hodgkin, 1988); it differs in that it contains a lower concentration of MgSO4 to prevent salt precipitation after autoclaving. - Sodium hypochlorite solution
5.4 parts distilled water
3 parts sodium hypochlorite (6%) or regular Bleach
1.6 parts 5 N NaOH
Prepare fresh in 15 ml or 50 ml polypropylene tube prior to use - Creating a platinum-wire worm pick
- Cut a 3-4 cm piece of 0.25 mm-diameter platinum wire
- Wrap a long Pasteur pipet in a paper towel and break the glass at the point where the tapering starts
- Heat the broken end of the Pasteur pipet in a Bunsen burner so that it is completely melted. While holding the platinum wire with forceps or pliers, embed the platinum wire in the melted end of the Pasteur pipet (generally while both are in the Bunsen burner flame)
- Remove from the flame and allow the glass to cool. The end of the platinum wire can be flattened and shaped as desired
- Cut a 3-4 cm piece of 0.25 mm-diameter platinum wire
- Tetracycline stock
- Dissolve 12.5 mg/ml tetracycline in 70% ethanol and 30% distilled water
- Filter with a 150 ml or 500 ml PES-membrane filter unit with a separate bottom container
- Wrap the bottom filter container with aluminum foil to protect the tetracycline solution from light. Store at -20 °C for over six months
- Dissolve 12.5 mg/ml tetracycline in 70% ethanol and 30% distilled water
- Chloramphenicol stock
Dissolve 34 mg/ml chloramphenicol in 100% ethanol
Store at -20 °C for over six months - Kanamycin stock
Dissolve 25 mg/ml kanamycin in distilled water
Filter sterilize and store at -20 °C for over six months - Cholesterol stock
Dissolve 10 mg/ml cholesterol in 100% ethanol
Store at room temperature for over twelve months - Antibiotic-enriched PBS with heat-killed bacteria
Sterile PBS supplemented with:
2% penicillin/streptomycin (from 100x stock)
25 µg/ml tetracycline (from 12.5 mg/ml stock)
34 µg/ml chloramphenicol (from 34 mg/ml stock)
50 µg/ml kanamycin (from 50 mg/ml stock)
0.2% normocin (from 500x stock)
10 µg/ml cholesterol (from 10 mg/ml stock)
Heat-killed bacteria are added to make the solution a lightly-cloudy white color (~50-200 µl of the heat-killed bacteria/2.5 ml PBS solution) - Hemin chloride stock
Prepare a fresh solution of 2 mM hemin chloride by dissolving 13 mg of hemin chloride in 9.8 ml water plus 0.2 ml 5 N NaOH. Prepare fresh each day - Stock of Hoechst 33342, calcein-AM, and ethidium homodimer
- Prepare a 1.6 mM stock of ethidium homodimer in DMSO. Shield from light and store at -80 °C
- Prepare a 10 mg/ml stock of Hoechst 33342 in distilled water. Shield from light and store at -80 °C
- Prepare a 1.6 mM stock of calcein-AM in distilled water. Shield from light and store at -80 °C
- To create 40 µl of stock solution, mix 1 µl ethidium homodimer stock solution (40 µg/ml final concentration); 8 µl Hoechst 33342 stock solution (2 mg/ml final concentration); 10 µl calcein-AM stock solution (0.4 mg/ml final concentration); 3 µl DMSO; and 18 µl distilled water. The stock solution can be stored shielded from light at -20 °C or -80 °C
Note: If one or more stain is too faint or bright, the concentration can be adjusted in the stock solution, with concomitant decreases or increases in the stain’s solvent (DMSO or water).
- Prepare a 1.6 mM stock of ethidium homodimer in DMSO. Shield from light and store at -80 °C
Acknowledgments
This protocol has been adapted from Chaudhari et al., 2016. Some C. elegans strains used to create strain ET507 were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by grants from NIH/NIGMS (R01GM074212) and NSF (MCB-1138454) to E.T.K.
References
- Bertani, G. (1951). Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62(3): 293-300.
- Chaudhari, S. N., Mukherjee, M., Vagasi, A. S., Bi, G., Rahman, M. M., Nguyen, C. Q., Paul, L., Selhub, J. and Kipreos, E. T. (2016). Bacterial folates provide an exogenous signal for C. elegans germline stem cell proliferation. Dev Cell 38(1): 33-46.
- Hansen, D. and Schedl, T. (2013). Stem cell proliferation versus meiotic fate decision in Caenorhabditis elegans. Adv Exp Med Biol 757: 71-99.
- Kimble, J. and Seidel, H. (2013). C. elegans germline stem cells and their niche. StemBook. Harvard Stem Cell Institute. 1-12.
- Strange, K., Christensen, M. and Morrison, R. (2007). Primary culture of Caenorhabditis elegans developing embryo cells for electrophysiological, cell biological and molecular studies. Nat Protoc 2(4): 1003-1012.
- Sulston, J. and Hodgkin, J. (1988). The Nematode Caenorhabditis elegans. In: Wood, W. B. (Ed). Cold Spring Harbor Laboratory 587-606.
- Zhang, S. and Kuhn, J. R. (2013). Cell isolation and culture. WormBook 1-39.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Vagasi, A. S., Rahman, M. M., Chaudhari, S. N. and Kipreos, E. T. (2017). Primary Culture System for Germ Cells from Caenorhabditis elegans Tumorous Germline Mutants. Bio-protocol 7(15): e2424. DOI: 10.21769/BioProtoc.2424.
Category
Stem Cell > Germ cell > In vitro culture
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link