- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Separation of Plant 6-Phosphogluconate Dehydrogenase (6PGDH) Isoforms by Non-denaturing Gel Electrophoresis
Published: Vol 7, Iss 14, Jul 20, 2017 DOI: 10.21769/BioProtoc.2399 Views: 9397
Reviewed by: Amit DeyAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1475 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3210 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1825 Views
Abstract
6-Phosphogluconate dehydrogenase (6PGDH; EC 1.1.1.44) catalyzes the third and irreversible reaction of the pentose phosphate pathway (PPP). It carries out the oxidative decarboxylation of the 6-phosphogluconate to yield ribulose-5-phosphate, carbon dioxide and NADPH. In higher plants, 6PGDH has several subcellular localizations including cytosol, chloroplast, mitochondria and peroxisomes (Corpas et al., 1998; Krepinsky et al., 2001; Mateos et al., 2009; Fernández-Fernández and Corpas, 2016; Hölscher et al., 2016). Using Arabidopsis thaliana as plant model and sweet pepper (Capsicum annuum L.) fruits as a plant with agronomical interest, this protocol illustrates how to prepare the plant extracts for the separation of the potential 6PGDH isoforms by electrophoresis on 6% polyacrylamide non-denaturing gels. Thus, this method allows detecting three 6PGDH isoforms in Arabidopsis seedlings and two 6PGDH isoforms in sweet pepper fruits.
Keywords: NADPHBackground
Non-denaturing gel electrophoresis is a powerful technique that allows separating native proteins. Their mobility depends of protein size, shape and net charge. In these analytical conditions, the protein preserves its activity and in combination with a specific staining method, it allows to separate the presence of potential isoforms. This approach has been widely used in the case of the family of superoxide dismutases. However, to our knowledge, there are not many papers that analyze the presence of different isoforms of 6PGDH activity in plant tissues (Corpas et al., 1998; Mateos et al., 2009). This straight method may be very useful for researchers working with plant 6PGDHs.
Materials and Reagents
- 10-cm-diameter Petri dishes
- Arabidopsis thaliana ecotype Columbia seeds (originally obtained from NASC, Nottingham Arabidopsis Stock Center)
- Sweet green pepper fruits were provided by Syngenta Seeds S.A. (El Ejido, Spain)
- 70% (v/v) ethanol
- 0.1% (w/v) SDS
- Commercial Bleach
- Murashige and Skoog medium (Sigma-Aldrich, catalog number: M5524 )
- Phyto-agar (Duchefa Biochemie, catalog number: P1001 )
- Sucrose (Sigma-Aldrich, catalog number: 84097 )
- Bio-Rad Protein Assay (Bio-Rad Laboratories, catalog number: 5000006 )
- Bovine serum albumin (BSA) Fraction V (Roche Diagnostics, catalog number: 10735078001 )
- Tris (AMRESCO, catalog number: 0497 )
- Ethylenediaminetetraacetic acid, disodium salt, dihydrate (Na2-EDTA) (Sigma-Aldrich, catalog number: E5134 )
- Triton X-100 (AMRESCO, catalog number: 0694 )
- Glycerol (AMRESCO, catalog number: E520 )
- Dithiothreitol (DTT) (Roche Diagnostics, catalog number: 10708984001 )
- 30% acrylamide/Bis solution, 19:1 (Bio-Rad Laboratories, catalog number: 1610154 )
- 4x gel buffer
- Glycine (AMRESCO, catalog number: 0167 )
- Ammonium persulfate
- TEMED
- β-Nicotinamide adenine dinucleotide phosphate, sodium salt, hydrate (NADP) (Sigma-Aldrich, catalog number: N0505 )
- Magnesium chloride, hexahydrate (MgCl2·7H2O) (EMD Millipore, catalog number: 442615 )
- Nitroblue tetrazolium (NBT) (AMRESCO, catalog number: 0329-1G )
- Phenazine methosulfate (PMS) (Sigma-Aldrich, catalog number: P9625 )
- 6-Phosphogluconic acid, trisodium salt (6PG) (Sigma-Aldrich, catalog number: P7877 )
- Glacial acetic acid (Fisher Scientific)
- Grinding buffer (see Recipes)
- Non-denaturing polyacrylamide gel electrophoresis (PAGE) on 6% acrylamide gel (see Recipes)
- Staining solution (see Recipes)
- Stop solution (see Recipes)
Equipment
- Plant Growth cabinet (Panasonic Biomedical, model: MLR-352-PE )
- Porcelain mortar and pestle
- Refrigerated centrifuge
- Vertical Slab gels Electrophoresis System (Bio-Rad Laboratories, model: Mini-PROTEAN® Electrophoresis Cell )
Software
- ImageJ program (https://imagej.nih.gov/ij/)
Procedure
- Plant extracts
- Surface-sterilize Arabidopsis thaliana ecotype Columbia seeds for 5 min in 70% (v/v) ethanol containing 0.1% (w/v) SDS at room temperature and with a slight shaking. Then, place for 20 min in sterile water containing 20% (w/v) commercial Bleach and 0.1% (w/v) SDS and then wash four times in sterile water.
- Grow seeds on Petri plates over commercial Murashige and Skoog medium (Sigma-Aldrich) with a pH of 5.5, containing 1% (w/v) sucrose and 0.8% (w/v) phyto-agar (Corpas and Barroso, 2014). Then, keep the seedlings for 14 days with a photoperiod of 16 h (light intensity of 100 μE m-2 sec-1) and 8 h dark, with a temperature of 22 °C and 18 °C, respectively.
- Sweet green pepper fruits were provided by Syngenta Seeds S.A. (El Ejido, Spain) from plants grown in experimental greenhouses.
- Homogenize plant samples in a mortar and pestle with grinding buffer (see Recipes) in a ratio 1:3 (w/v) for Arabidopsis and ratio 1:1 for pepper fruits. Perform these operations at 0-4 °C (on ice).
- Centrifuge extracts at 27,000 x g at 4 °C for 20 min.
- Use the supernatants for the enzymatic assays. Determine protein concentration with the Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, CA) using bovine serum albumin as standard.
- Surface-sterilize Arabidopsis thaliana ecotype Columbia seeds for 5 min in 70% (v/v) ethanol containing 0.1% (w/v) SDS at room temperature and with a slight shaking. Then, place for 20 min in sterile water containing 20% (w/v) commercial Bleach and 0.1% (w/v) SDS and then wash four times in sterile water.
- 6PGDH activity staining
- Separate plant proteins by non-denaturing polyacrylamide gel electrophoresis (PAGE) on 6% acrylamide gels at 4-7 °C. Usually, it takes around 2 h and 30 min.
- After electrophoresis, incubate gels in staining solution (see Recipes) at room temperature (RT). Keep in the dark with shaking until blue-purple bands appear over a colorless background. It takes around 15 min (Figure 1).
- Remove the staining solution and add stop solution (see Recipes) to cover gels.
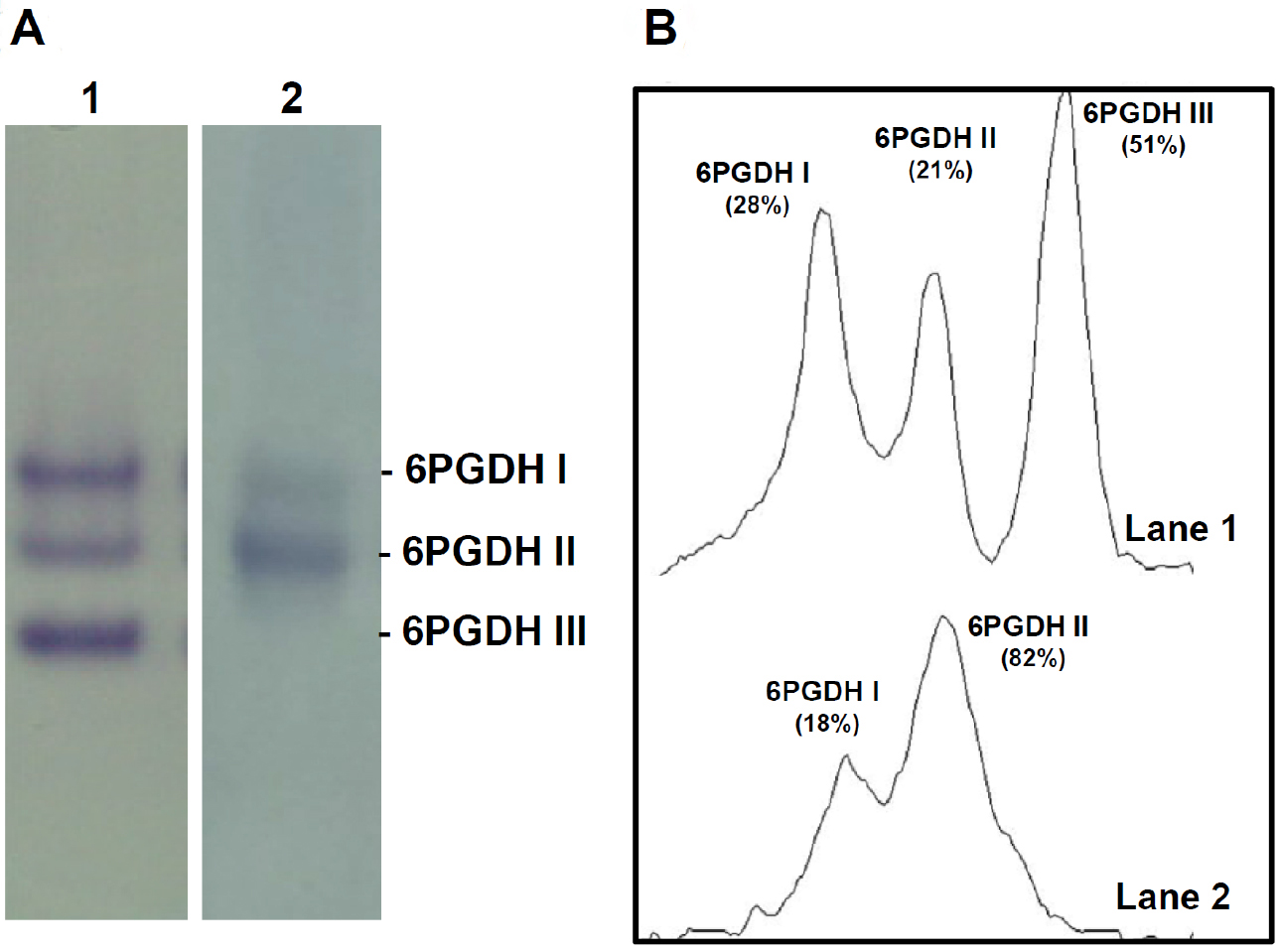
Figure 1. Analysis of 6-phosphogluconate dehydrogenase (6PGDH) isoforms by non-denaturing gel electrophoresis. A. 6PGDH isoforms activity were separated by native PAGE (6%) and identified by activity staining. Lane 1, Arabidopsis thaliana 14-day-old seedlings. Lane 2, sweet green pepper fruits. B. Densitometric scans of 6PGDH isoforms and its relative quantification (%) made by the ImageJ program.
- Separate plant proteins by non-denaturing polyacrylamide gel electrophoresis (PAGE) on 6% acrylamide gels at 4-7 °C. Usually, it takes around 2 h and 30 min.
Data analysis
Quantification of the activity of each 6PGDH isoform can be done by densitometric analysis (Figure 1B), for example, using the ImageJ program.
Recipes
- Grinding buffer
50 mM Tris-HCl, pH 7.8 containing 0.1 mM EDTA
0.2% (v/v) Triton X-100
10% (v/v) glycerol
2 mM DTT - Non-denaturing polyacrylamide gel electrophoresis (PAGE) on 6% acrylamide gel
30% acrylamide/Bis solution, 19:1
4x gel buffer (1.5 M Tris-HCl, pH 8.9)
Electrode buffer (38 mM glycine plus 5 mM Tris, pH 8.2)
10% (w/v) ammonium persulfate (AP)
TEMED
The follow table shows the needed volume of each reagent to prepare two 1.5 mm thick gels to be used in a Bio-Rad Mini-PROTEAN Electrophoresis System
*Note: Added right before each use. The mix of the reagents should be done at 4 °C (on ice).
The standard running conditions are 15 mA per gel for 30 min and then increase to 25 mA per gel for approximately 90 min. The electrophoresis should be run in a cold room (5-7 °C) - Staining solution
50 mM Tris-HCl, pH 7.6 containing 0.8 mM NADP
5 mM EDTA
2 mM MgCl2
0.24 mM NBT
65 mM PMS
10 mM 6PG
Note: Freshly prepared before use and protected from the light. - Stop solution
7% (v/v) acetic acid
Acknowledgments
This work has been supported by the ERDF-cofinanced grant AGL2015-65104-P from the Ministry of Economy and Competitiveness, Spain.
References
- Corpas, F. J. and Barroso, J. B. (2014). Peroxynitrite (ONOO-) is endogenously produced in Arabidopsis peroxisomes and is overproduced under cadmium stress. Ann Bot 113(1): 87-96.
- Corpas, F. J., Barroso, J. B., Sandalio, L. M., Distefano, S., Palma, J. M., Lupiáñez, J. A. and del Río, L. A. (1998). A dehydrogenase-mediated recycling system of NADPH in plant peroxisomes. Biochem J 330 (Pt 2): 777-784.
- Fernández-Fernández, A. D. and Corpas, F. J. (2016). In silico analysis of Arabidopsis thaliana peroxisomal 6-phosphogluconate dehydrogenase. Scientifica (Cairo) 2016: 3482760.
- Hölscher, C., Lutterbey, M. C., Lansing, H., Meyer, T., Fischer, K. and von Schaewen, A. (2016). Defects in peroxisomal 6-phosphogluconate dehydrogenase isoform PGD2 prevent gametophytic interaction in Arabidopsis thaliana. Plant Physiol 171(1): 192-205.
- Krepinsky, K., Plaumann, M., Martin, W. and Schnarrenberger, C. (2001). Purification and cloning of chloroplast 6-phosphogluconate dehydrogenase from spinach. Cyanobacterial genes for chloroplast and cytosolic isoenzymes encoded in eukaryotic chromosomes. Eur J Biochem 268(9): 2678-2686.
- Mateos, R. M., Bonilla-Valverde, D., del Río, L. A., Palma, J. M. and Corpas, F. J. (2009). NADP-dehydrogenases from pepper fruits: effect of maturation. Physiol Plant 135(2): 130-139.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Corpas, F. J., de Freitas-Silva, L., García-Carbonero, N., Contreras, A., Terán, F., Ruíz-Torres, C. and Palma, J. M. (2017). Separation of Plant 6-Phosphogluconate Dehydrogenase (6PGDH) Isoforms by Non-denaturing Gel Electrophoresis. Bio-protocol 7(14): e2399. DOI: 10.21769/BioProtoc.2399.
Category
Plant Science > Plant biochemistry > Protein > Activity
Plant Science > Plant physiology > Metabolism
Biochemistry > Protein > Electrophoresis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











