- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Non-invasive Protocol for Kinematic Monitoring of Root Growth under Infrared Light
(*contributed equally to this work) Published: Vol 7, Iss 14, Jul 20, 2017 DOI: 10.21769/BioProtoc.2390 Views: 9616
Reviewed by: Igor Cesarino Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Live Imaging of the Shoot Apical Meristem of Intact, Soil-Grown, Flowering Arabidopsis Plants
Gabriele Bradamante
Jun 20, 2024 2286 Views

Micrografting Technique of Hevea brasiliensis In Vitro Plantlets
Florence Dessailly [...] Julie Leclercq
Feb 20, 2025 1491 Views

Enzymatic Starch Quantification in Developing Flower Primordia of Sweet Cherry
Nestor Santolaria [...] Afif Hedhly
Apr 5, 2025 1896 Views
Abstract
Phenotyping the dynamics of root responses to environmental cues is necessary to understand plant acclimation to their environment. Continuous monitoring of root growth is challenging because roots normally grow belowground and are very sensitive to their growth environment. This protocol combines infrared imaging with hydroponic cultivation for kinematic analyses. It allows continuous imaging at fine spatiotemporal resolution and disturbs roots minimally. Examples are provided of how the procedure and materials can be adapted for 3D monitoring and of how environmental stress may be manipulated for experimental purposes.
Keywords: HydroponicsBackground
The use of kinematic analyses for the monitoring of growth and tropisms in plants dates back to the end of the 19th century, with early studies from Julius von Sachs and Whilhelm Pfeffer. The widespread use of photography during the 20th century led to easier and continuous monitoring through ‘streak photography’ (List, 1969; Erickson and Silk, 1980). In the 90s, new digital cameras and informatics tools enabled the development of automatic tracking algorithms used for particle image velocimetry. RootFlowRT (Van der Weele et al., 2003), Kineroot (Basu et al., 2007), RootTrace (French et al., 2009), GrowthTracer (Iwamoto et al., 2013) and Kymorod (Bastien et al., 2016) are among many recent examples of software dedicated to the monitoring of root growth. However, all particle image velocimetry methods rely on the use of identifiable image texture patterns in each successive picture. Historically, these patterns were marked on the root using ink or graphite particles (see Sharp et al., 1988 and Merret et al., 2010 for examples). Two difficulties arise from this approach: Firstly, the markings spread during organ (root) growth, inducing loss of resolution after a few hours. Secondly, the physical marking process may stress the organ due to some combination of exposure to small forces, to light or to temperature changes, or to slight drying of the organ surface. In roots such handling effects may cause temporary slowing of growth for a period of minutes to hours. The use of infrared light (840-850 nm) has the double benefit of not influencing root growth, whilst also generating image texture patterns readily tracked using particle image velocimetry.
Materials and Reagents
- Clear flexible tubing (VWR, catalog number: 228-0708 )
- Plastic sealing tape (Terostat VII, Teroson) (Rubans de Normandie, catalog number: 7TE )
- Growing roots of hydroponically-grown plants. Adventitious roots grow on poplar woody cuttings partially immersed in Hoagland half strength nutrient solution (Sigma-Aldrich, catalog number: H2395 )
Note: The present protocol was initially designed for monitoring the growth of a plagiotropic poplar root (Bizet et al., 2015). The protocol is adaptable for any plant species that tolerates hydroponic or in vitro cultivation (see step D3).
Equipment
- Transparent Plexiglass® growth monitoring chamber (specially built, size 12 cm long x 5 cm large x 6 cm height), with holes on one side for nutrient solution inflow and outflow (Figure 1)

Figure 1. Custom made transparent Plexiglass® growth monitoring chamber - Water pump (e.g., a small aquarium pump, for instance, Newa, model: NJ600 , aquarium shop)
- Temperature-controlled dark room (range 21-23 °C)
Note: The whole system requires about 1 m2 of laboratory working space. - Air pump (e.g., an aquarium pump with airstone, for instance JBL, model: Prosilent a300 , aquarium shop)
- Stone diffuser (e.g., JBL, model: Prosilent Aeras Micro S2 , aquarium shop)
- Small plastic reservoir tank (buffer tank, few litres capacity, size 12 x 20 x 40 cm3)
Note: A plastic bucket would suit. - Modified digital camera (see Procedure A, for how to remove the infrared filter)
Note: Most reflex camera should work fine as long as it is possible to do infrared conversion and to computer-control them using manufacturer’s software or free software such as Digicamcontrol (list of supported cameras: http://digicamcontrol.com/cameras). Light sensitivity and captor size should also be considered for best image quality. - Infrared lamp (840-850 nm) (Pearl, catalog number: KT4243-907 )
- Extension tube (Kenko, AF 12/20/36 mm for Nikon)
- Optical rail equipped with a translation stage (Edmund Optics, catalog number: 59-263 )
- Knuckle with knob and thread adaptor (Edmund Optics, catalog numbers: 53-887 and 58-988 )
- Macro objective (Nikkor 60/2.8 D ASF)
Software
- Software for image analysis (e.g., Kineroot from Basu et al., 2007)
- Fiji (https://fiji.sc)
- Rawtherapee (http://rawtherapee.com)
- Digicamcontrol (http://digicamcontrol.com)
Procedure
- Digital camera modification for infrared detection
The use of infrared light requires a modified digital camera able to detect wavelengths above 700 nm. Fortunately, the technology behind charged coupled device (CCD) sensors used in digital cameras can detect light up to more than 1,000 nm. The sensitivity of the sensor is however limited by a low-pass filter blocking infrared light above 720-750 nm, depending on manufacturer. Thus, to use a digital camera for infrared imaging, removal of the low-pass filter is necessary−its replacement by a colourless glass filter (e.g., Schott WG280) is desirable to protect against dust particles on the sensor. Filter replacement is widely used for astronomical photography, and tutorials showing how to remove the low-pass filter without damaging the camera CCD sensor are readily available on the internet (https://www.lifepixel.com/tutorials/infrared-diy-tutorials), with specialist companies also offering this service (e.g., above mentioned LifePixel or http://www.eosforastro.com). Figure 2 shows examples of roots from two different species filmed with a modified camera under infrared light.
Note: The choice of a high-resolution digital camera with good quality (e.g., macro) lens is very important to avoid optical aberration (distortion) of images. This is essential to ensure that tracking of organ growth is accurate across the entire image frame. To check for lens aberrations an accurately drawn square grid should be photographed and the degree of aberration checked–it is possible to digitally correct for aberration, but it is far better to select the camera optics to avoid these problems. The grid can also be used to calibrate the growth measurements from pixels into distance measurements.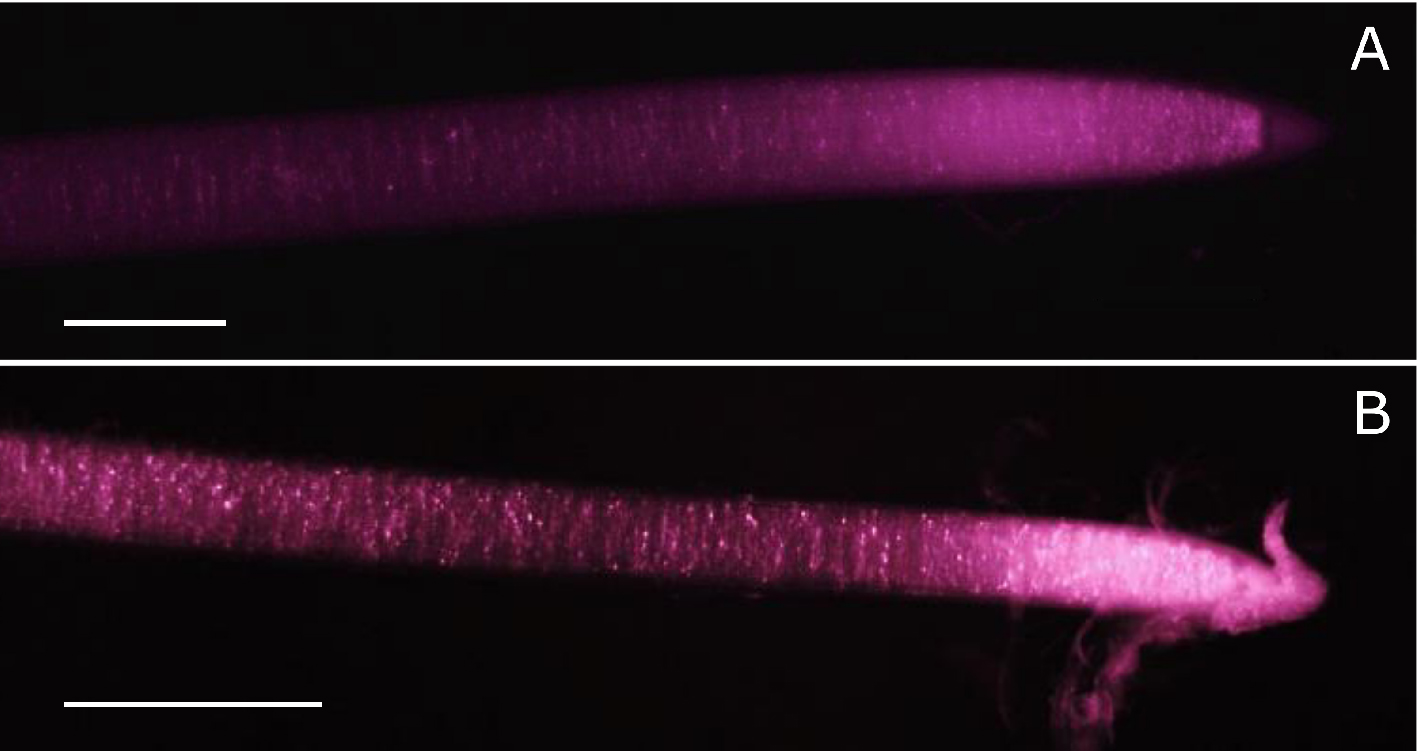
Figure 2. Examples of root apices under infrared light for maize (A) and poplar (B). Pictures were taken with a defiltered Nikon D5200 mounted with a macro objective (Nikkor 60 mm). Scale bars stand for 1 mm. Picture for maize was taken from Bizet et al. (2015).Note: Subsequent steps are given for plagiotropic roots obtained from poplar cuttings cultivated in hydroponics. Procedure can be adapted for gravitropic roots from various species using an adapted growth monitoring chamber and material to easily attach the plant.
- Root growth monitoring under near-infrared light
- Preparation of the growth monitoring chamber
- Fill the buffer tank with approx. ten times the volume of the growth monitoring chamber with nutrient solution and place it below the growth monitoring chamber.
- Set up circulation of the nutrient solution between the growth monitoring chamber and the buffer tank using a water pump and flexible tubing: add an inflow from the pump to the bottom of the growth monitoring chamber and a passive outflow from the top of the growth monitoring chamber to the buffer tank (Figure 3A). Flow rate should be small, 2.5 to 3 L h-1.
- Ensure sealing of the nutrient circulation system using plastic sealing tape.
- Add a source of dissolved oxygen inflow in the buffer tank using an air pump, airline tubing and an air stone diffuser (Figure 3B).
Note: The source of dissolved oxygen should not be added in the growth monitoring chamber to prevent shaking of the root by air bubbles during the monitoring.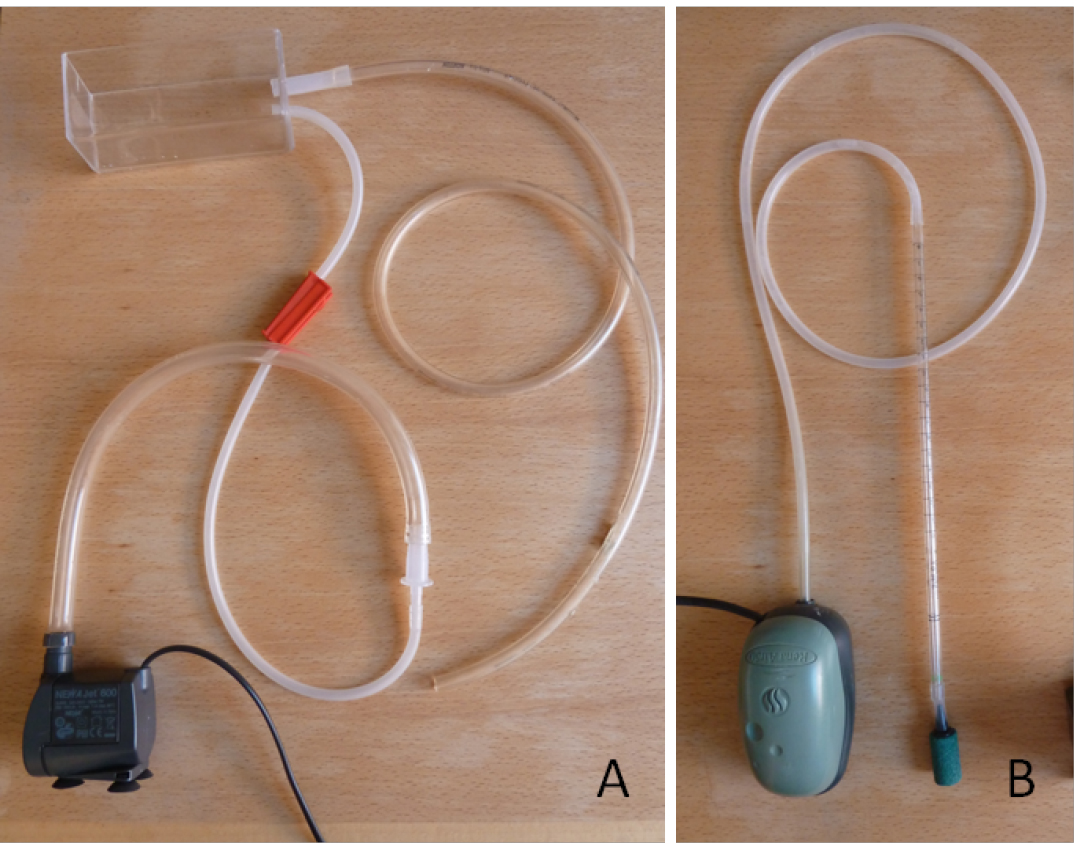
Figure 3. Nutrient solution circuit from the tank to the growth monitoring chamber and back to the tank (A) and the aeration system (B)
Note: All subsequent steps should be done in a dark room.
- Fill the buffer tank with approx. ten times the volume of the growth monitoring chamber with nutrient solution and place it below the growth monitoring chamber.
- Gently place a root in the growth monitoring chamber and attach the plant, e.g., for poplar roots: clamp the cutting.
- Add an infrared lamp in front of the root apex.
Note: A low incidence angle between the root axis and the lamp allow better visualization as long as the lamp is not oriented toward the camera. - Place the defiltered camera for a side view of the root. The camera is attached to a micrometric translation stage mounted on an optical rail (Figure 4).
Note: The camera should be equipped with a macro objective and extension tubes can be added to obtain higher magnification while keeping a close minimum focusing distance.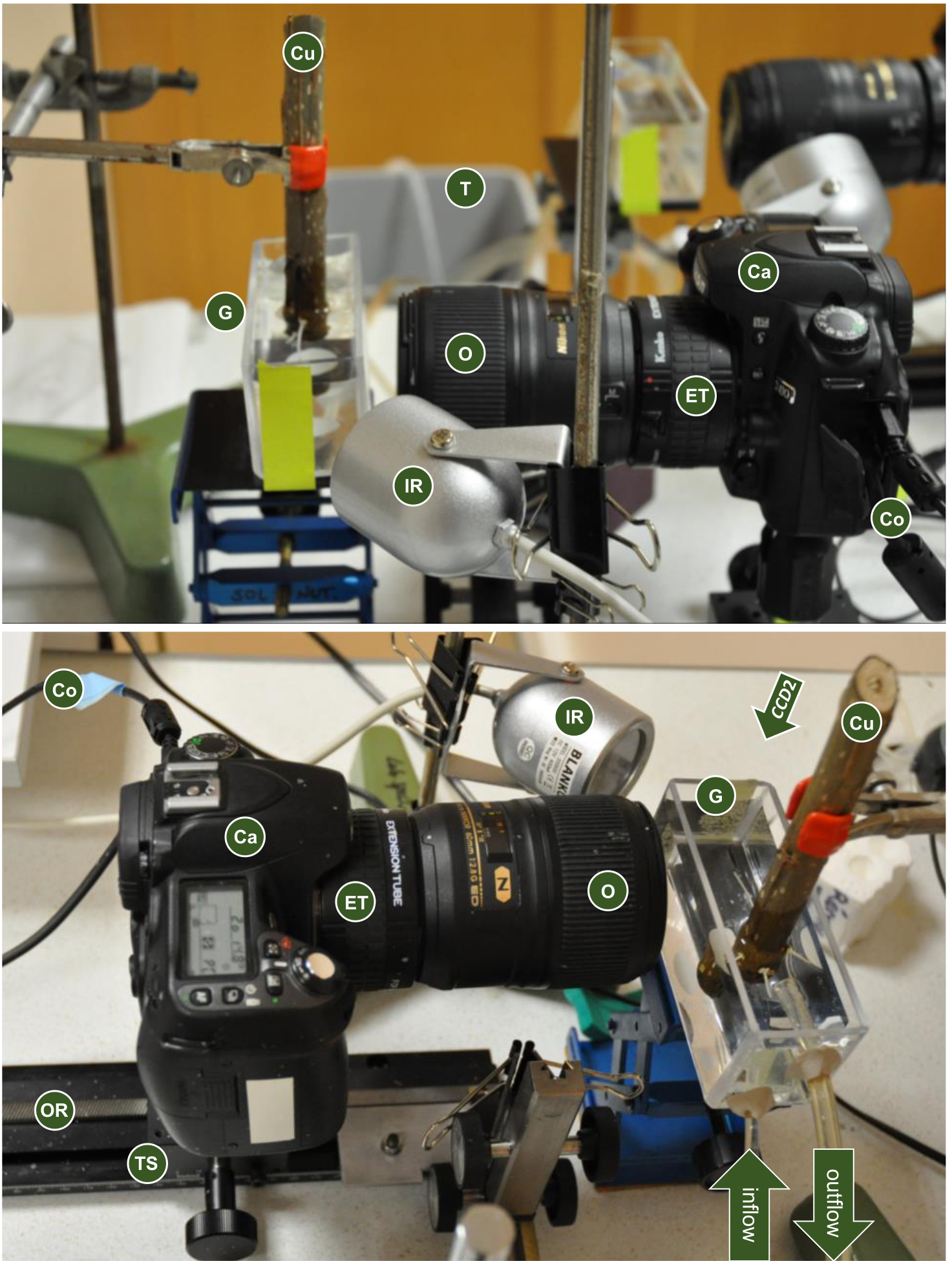
Figure 4. Components of the root growth monitoring system. Note the presence of the buffer tank (T) connected to the growth monitoring chamber (G) in the background of the top picture. Top picture also shows duplication of the system for parallel monitoring (top-right corner). CCD2 arrow indicates the possible position for a second top-view camera (see step D1). Ca: camera; Co: connection to computer; Cu: cutting; ET: extension tube; G: growth monitoring chamber; IR: infrared lamp; O: macro objective; OR: optical rail; T: buffer tank; TS: translation stage. - Set up the focus on the root using the translation stage.
Notes:- Video streaming on a computer can greatly help obtain a precise focus. Keep the focusing distance on the macro objective at minimum and only use the translation stage during this step to maximize the magnification.
- For calibration purposes, take a picture of a ruler (either at the beginning or the end of the monitoring). The scale is valid as long as the focal length and distance between the objective and the root/ruler is conserved (meaning the magnification is identical).
- For long term experiment (several days), focus adjustment and framing may be necessary once or twice a day as the root may go out of the camera depth of field and out of the frame.
- Video streaming on a computer can greatly help obtain a precise focus. Keep the focusing distance on the macro objective at minimum and only use the translation stage during this step to maximize the magnification.
- Launch the intervalometer of the camera. Selecting intervals from 3 min to 30 min between images allows capture of growth at varying resolutions.
Note: Most cameras can be computer-controlled using manufacturers’ software or free software such as Digicamcontrol (http://digicamcontrol.com). - At the end of the experiment, wash all the materials that were in contact with the nutrient solution.
Note: Be careful not to scratch the growth monitoring chamber as this degrades image quality and makes organ tracking difficult or impossible.
- Preparation of the growth monitoring chamber
- Image processing
Depending on the temporal resolution desired, many images may be processed during kinematic analysis. To optimize particle image velocimetry calculation time and resolution (e.g., small roots of Arabidopsis), subsequent steps of image pre-processing should be considered. These pre-processing steps can be done with free software that allows batch processing such as Fiji (https://fiji.sc) or Rawtherapee (http://rawtherapee.com), the latter being able to read raw picture format from various manufacturers (for example .nef for Nikon or .crw for Canon). We used batch processing of Rawtherapee (see the online help files for details), with the following steps:- Crop images to optimize calculation time through deletion of unused information.
- Adapt the black threshold to decrease background noise.
- Remove the presence of overexposed areas.
- Modify local contrasts to blur unused pixel scale details.
- Enhance patches of pixels which are more stable between consecutive pictures (Figure 5A).
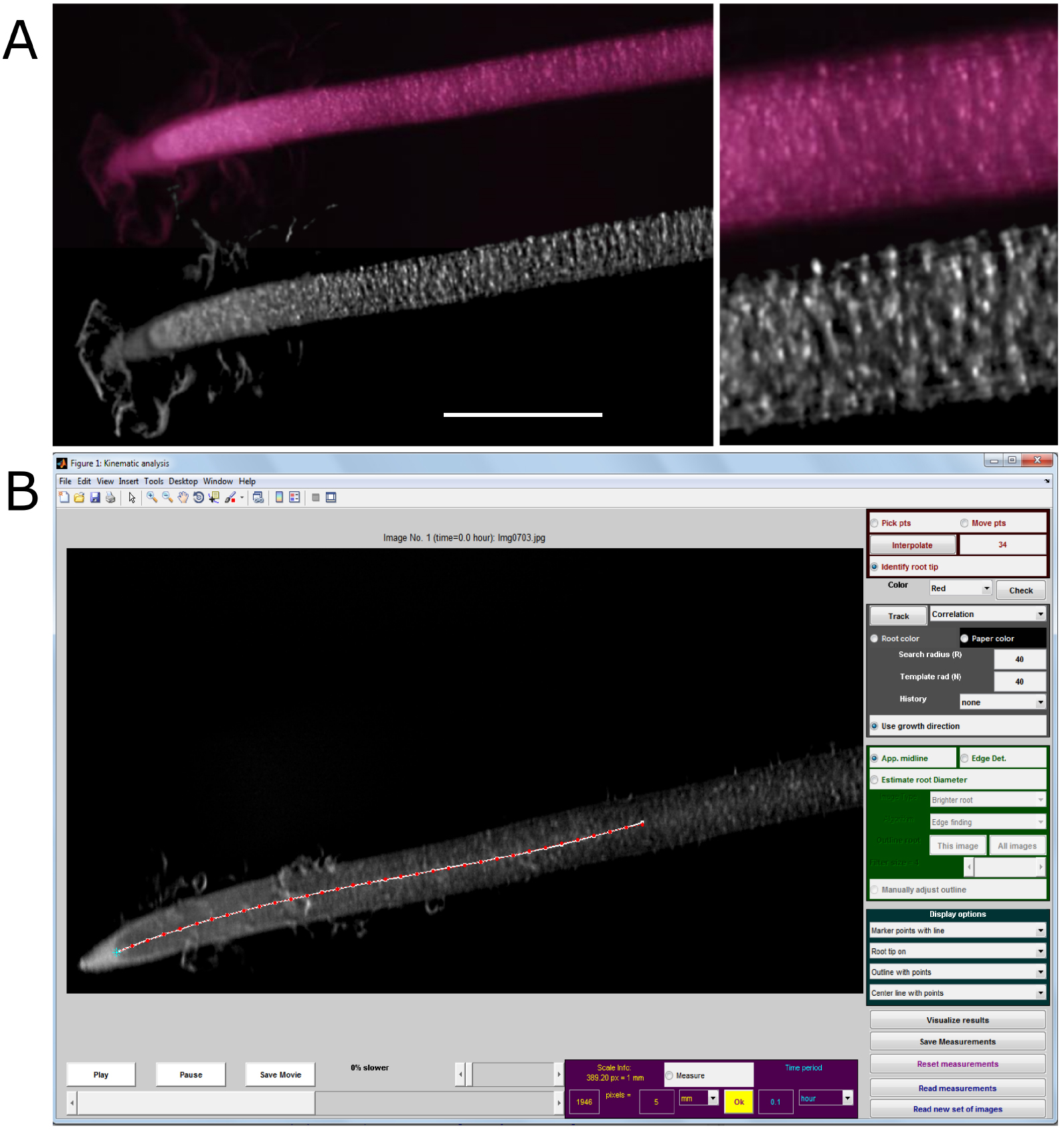
Figure 5. Use of pre-processed images for kinematic analysis on Kineroot software. A. Enhanced image texture patterns for kinematic analysis after image pre-processing (bottom) as compared to raw picture (top). Scale bar stands for 1 mm. B. Example of root monitoring with Kineroot. Red points are tracked along the root centerline by particle image velocimetry. Monitoring parameters can be seen in the different panels. Note that R and N parameters (both set here at 40 pixels) must be adapted to the spatial and temporal resolutions (389 pixels mm-1 and 6 min between consecutive pictures here). - Crop images to optimize calculation time through deletion of unused information.
- Flexibility of the growth monitoring system
- 3D monitoring of root shape and deformation: Adding a top view to the initial side view allows the reconstruction of the 3D shape of the root. Multiple cameras can be used simultaneously using Digicamcontrol (http://digicamcontrol.com). See Bizet et al. (2016) for detailed explanations about cameras position and how analysis from two points of view can be merged into a 3D reconstruction of the root apex (Figure 6).
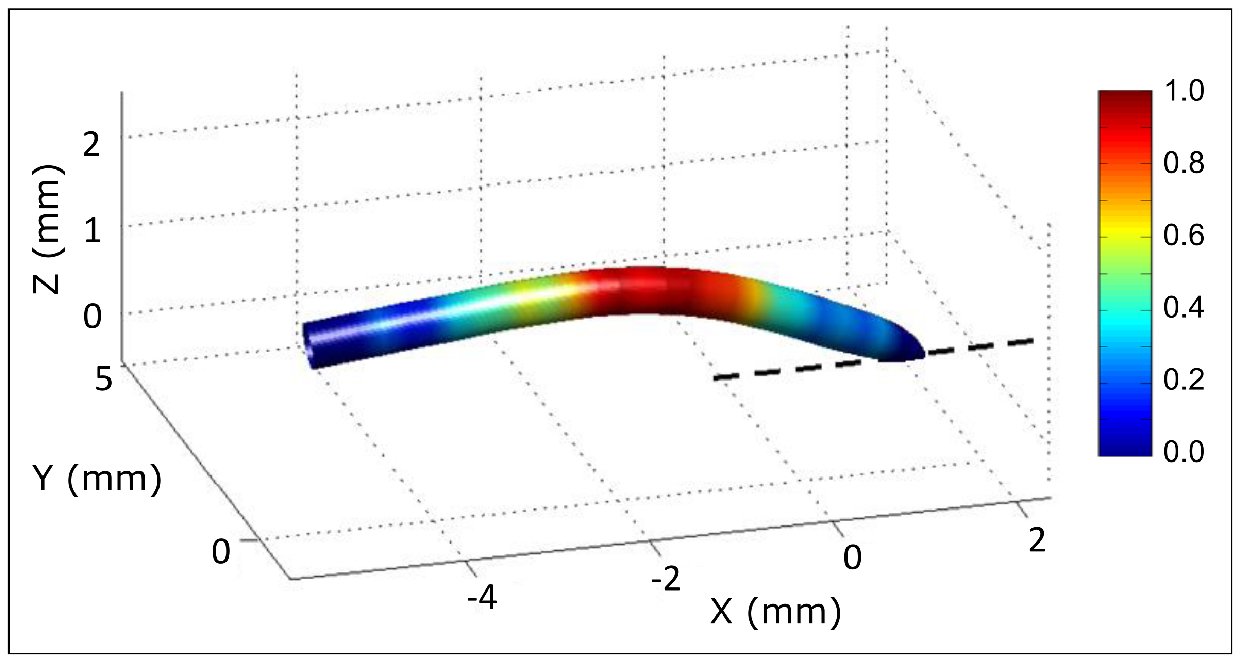
Figure 6. 3D reconstruction of a root from two orthogonal points of view. Colors indicate relative root curvature. - Stress management during continuous monitoring: The system for root growth monitoring described in Procedure B can be adapted to replace the nutrient solution by another solution whilst monitoring growth continually and without any manipulation of the root (see Bizet et al., 2015, for root exposure to osmotic stress using this system).
- Prior to the setup of the root in the growth monitoring chamber, connect a second buffer tank (tank b) in parallel with the initial buffer tank (tank a). Tank b contains the test solution and should be connected to the nutrient inflow using a ball valve (Thermo Fisher) (Figure 7). Check if solution temperature is identical in both tanks.
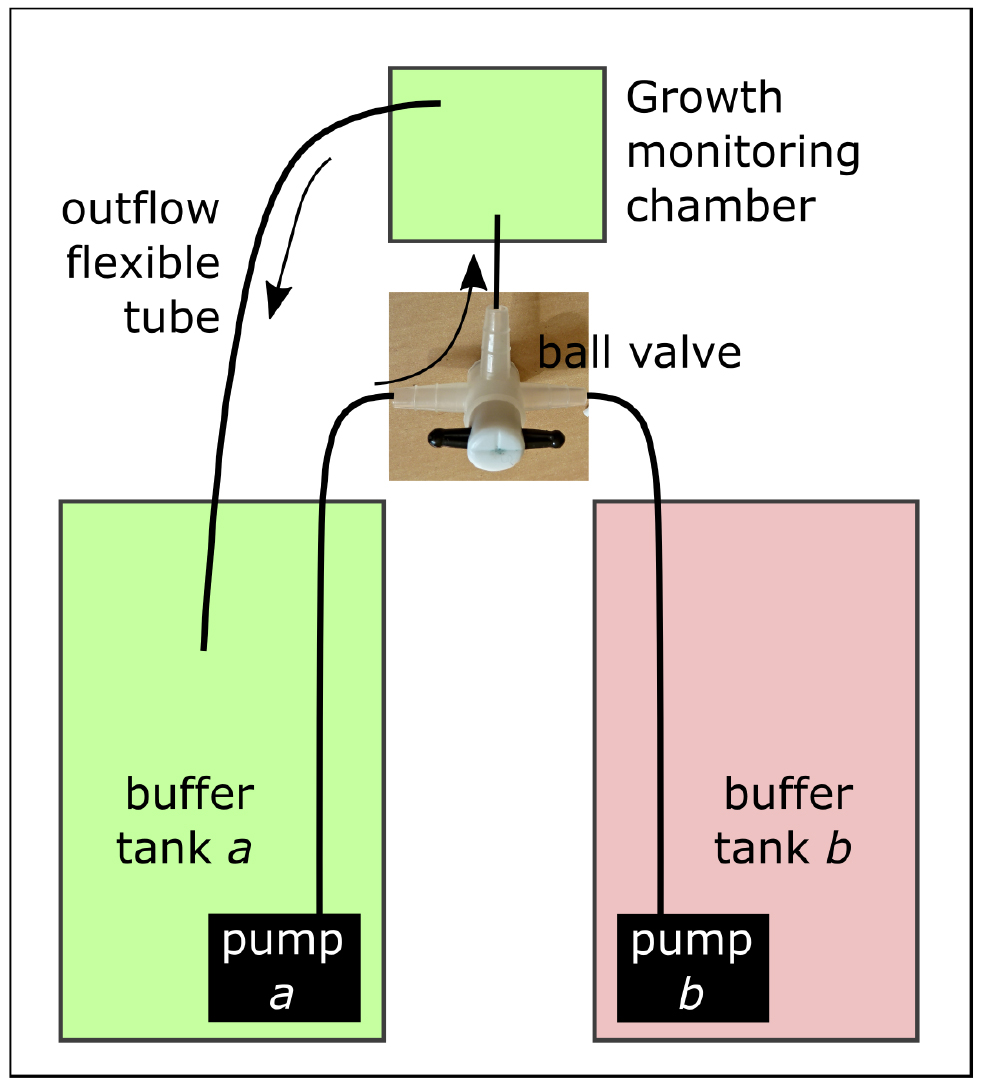
Figure 7. Set-up of the double tank with the ball valve for stress application - Close the ball valve from tank b and proceed from step B1 to B6.
- To change the circulating solution, turn on pump b, close the ball valve from tank a and open the ball valve from tank b.
- Wait five minutes for the solution in the growth monitoring chamber to be replaced and move the outflow flexible tube from buffer tank a to buffer tank b.
- At the end of the experiment, wash all the materials that were in contact with the nutrient solution.
- Prior to the setup of the root in the growth monitoring chamber, connect a second buffer tank (tank b) in parallel with the initial buffer tank (tank a). Tank b contains the test solution and should be connected to the nutrient inflow using a ball valve (Thermo Fisher) (Figure 7). Check if solution temperature is identical in both tanks.
- The protocol is flexible to suit the species and growth condition requirements. To monitor growth of gravitropic roots, use a vertical growth monitoring chamber. For in vitro plants, for which the root diameter is much finer, higher magnification may be obtained with more extension tubes on the camera, and the agar plate is directly used as the growth monitoring chamber.
- 3D monitoring of root shape and deformation: Adding a top view to the initial side view allows the reconstruction of the 3D shape of the root. Multiple cameras can be used simultaneously using Digicamcontrol (http://digicamcontrol.com). See Bizet et al. (2016) for detailed explanations about cameras position and how analysis from two points of view can be merged into a 3D reconstruction of the root apex (Figure 6).
Data analysis
Kinematic analyses by Kineroot provide values of velocity along the root apex, namely velocity profiles. Elemental elongation rate along the root apex is the first derivative of the velocity profile. Alternatively, a composite growth function dedicated to root velocity profiles can be fitted on Kineroot output (Peters and Baskin, 2006). See Bizet et al. (2015) for more details about the fitting procedure. Growth parameters such as root growth rate (that equals maximal velocity), growth zone length and maximal elemental elongation rate are extracted from these profiles (Figure 8). Parametric statistics can then be on to analyse these parameters, to compare individuals, or to examine the time course of the response to an environmental stress.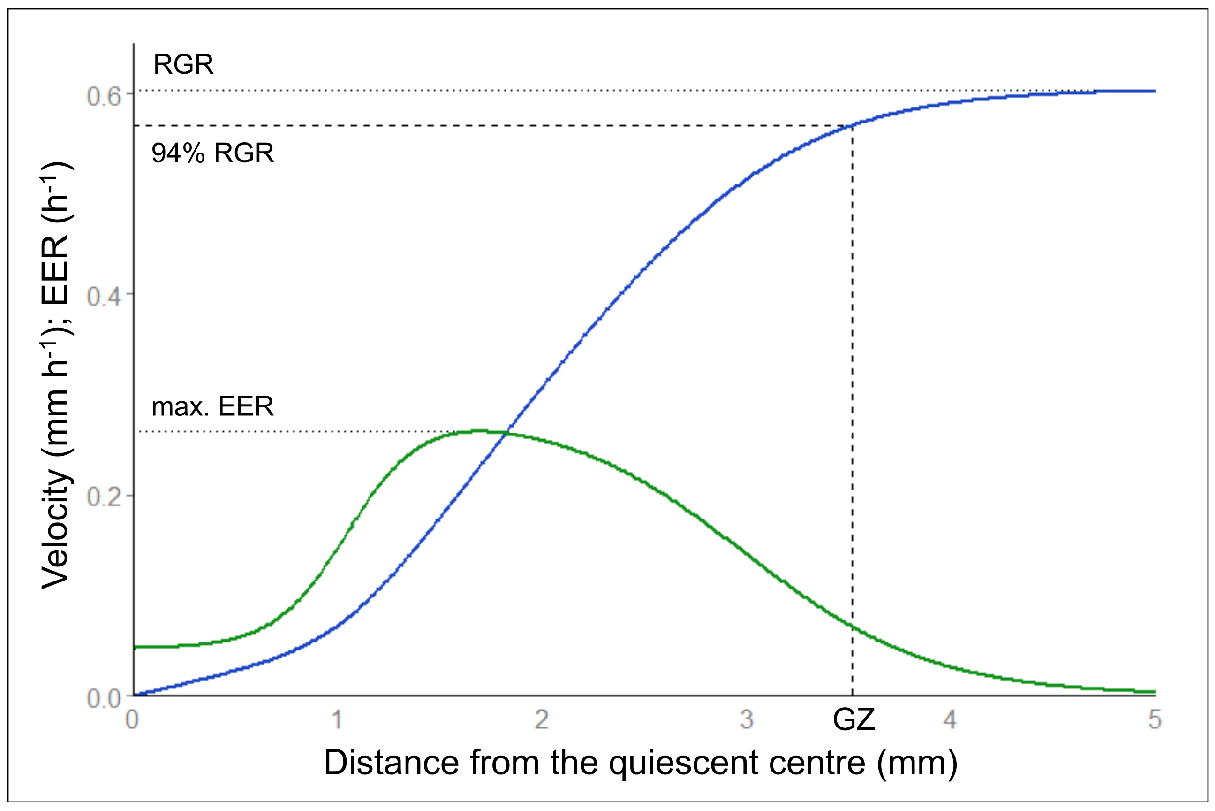
Figure 8. Velocity and elemental elongation rate profiles. Velocity profile (in blue) and elemental elongation rate (EER, in green) profile after fitting of the composite growth function from Peters and Baskin (2006). Among growth parameters, root growth rate (RGR) corresponds to the maximum of the velocity profile. Maximal EER corresponds to the maximal slope of the velocity profile i.e., the maximum of the first derivative. The growth zone length (GZ) is determined at 94% of the maximal velocity to avoid infinite values.
Notes
From our observations, growth rate is very variable between individual roots, varying up to 5 fold between minimal and maximal values. Experimental design should take into account this variability to maximise the power of statistical analyses.
Acknowledgments
FB was supported by a grant from the French National Ministry for Education and Research. The research has received funding from the French National Research Agency through the Laboratory of Excellence ARBRE (ANR-12-LABXARBRE-01) and from Agreenium’s International Research School. The James Hutton Institute receives funding from the Scottish Government. This protocol was adapted from Bizet et al. (2015) and Bizet et al. (2016).
References
- Bastien, R., Legland, D., Martin, M., Fregosi, L., Peaucelle, A., Douady, S., Moulia, B. and Hofte, H. (2016). KymoRod: a method for automated kinematic analysis of rod-shaped plant organs. Plant J 88(3): 468-475.
- Basu, P., Pal, A., Lynch, J. P. and Brown, K. M. (2007). A novel image-analysis technique for kinematic study of growth and curvature. Plant Physiol 145(2): 305-316.
- Bizet, F., Bengough, A. G., Hummel, I., Bogeat-Triboulot, M. B. and Dupuy, L. X. (2016). 3D deformation field in growing plant roots reveals both mechanical and biological responses to axial mechanical forces. J Exp Bot 67(19): 5605-5614.
- Bizet, F., Hummel, I. and Bogeat-Triboulot, M. B. (2015). Length and activity of the root apical meristem revealed in vivo by infrared imaging. J Exp Bot 66(5): 1387-1395.
- Erickson R. O. and Silk, W. K. (1980). The kinematics of plant growth. Sci Am 242(5): 134–151.
- French, A., Ubeda-Tomas, S., Holman, T. J., Bennett, M. J. and Pridmore, T. (2009). High-throughput quantification of root growth using a novel image-analysis tool. Plant Physiol 150(4): 1784-1795.
- Iwamoto, A., Kondo, E., Fujihashi, H. and Sugiyama, M. (2013). Kinematic study of root elongation in Arabidopsis thaliana with a novel image-analysis program. J Plant Res 126(1): 187-192.
- List, A., Jr. (1969). Transient growth responses of the primary roots of Zea mays. Planta 87(1-2): 1-19.
- Merret, R., Moulia, B., Hummel, I., Cohen, D., Dreyer, E. and Bogeat-Triboulot, M. B. (2010). Monitoring the regulation of gene expression in a growing organ using a fluid mechanics formalism. BMC Biol 8: 18.
- Peters, W. S. and Baskin, T. I. (2006). Tailor-made composite functions as tools in model choice: the case of sigmoidal vs bi-linear growth profiles. Plant Methods 2: 11.
- Sharp, R. E., Silk, W. K. and Hsiao, T. C. (1988). Growth of the maize primary root at low water potentials: I. Spatial distribution of expansive growth. Plant Physiol 87(1): 50-57.
- van der Weele, C. M., Jiang, H. S., Palaniappan, K. K., Ivanov, V. B., Palaniappan, K. and Baskin, T. I. (2003). A new algorithm for computational image analysis of deformable motion at high spatial and temporal resolution applied to root growth. Roughly uniform elongation in the meristem and also, after an abrupt acceleration, in the elongation zone. Plant Physiol 132(3): 1138-1148.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Bizet, F., Dupuy, L. X., Bengough, A. G., Peaucelle, A., Hummel, I. and Bogeat-Triboulot, M. (2017). Non-invasive Protocol for Kinematic Monitoring of Root Growth under Infrared Light. Bio-protocol 7(14): e2390. DOI: 10.21769/BioProtoc.2390.
Category
Plant Science > Plant developmental biology > General
Plant Science > Plant physiology > Plant growth
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









