- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Assay to Measure DNA Polymerase β Nucleotide Insertion Coupled with the DNA Ligation Reaction during Base Excision Repair
Published: Vol 7, Iss 12, Jun 20, 2017 DOI: 10.21769/BioProtoc.2341 Views: 7743
Reviewed by: Gal HaimovichOmar AkilAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Colocalizing Telomeres With PML or γH2AX Foci by IF-FISH in Mouse Brain Neurons
Anna Konopka
Nov 5, 2025 1515 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 496 Views

A Quantitative DNA Fiber Assay to Monitor Replication Fork Progression, Protection, and Restart
Debanjali Bhattacharya and Ganesh Nagaraju
Feb 5, 2026 225 Views
Abstract
We previously reported that oxidized nucleotide insertion by DNA polymerase β (pol β) can confound the DNA ligation step during base excision repair (BER) (Çağlayan et al., 2017). Here, we describe a method to investigate pol β nucleotide insertion coupled with DNA ligation, in the same reaction mixture including dGTP or 8-oxo-dGTP, pol β and DNA ligase I. This in vitro assay enables us to measure the products for correct vs. oxidized nucleotide insertion, DNA ligation, and ligation failure, i.e., abortive ligation products, as a function of reaction time. This protocol complements our previous publication and describes an efficient way to analyze activities of BER enzymes and the functional interaction between pol β and DNA ligase I in vitro.
Keywords: DNA base excision repairBackground
This protocol is to observe the last two steps of the BER pathway: nucleotide insertion by pol β and DNA ligation by ligase I. Using this protocol, one measures both reactions as a function of time of incubation in the same reaction mixture in vitro. The original entity was to analyze BER enzymes pol β and DNA ligase I on the single-nucleotide gapped DNA substrate that mimics a BER intermediate. These BER enzymes bind to and function on this BER intermediate. The DNA substrate used in this protocol includes a fluorescent label at both 5’- and 3’-ends that enables us to observe single-nucleotide insertion and DNA ligation of the DNA substrate. The pol β nucleotide insertion coupled with DNA ligation mimics the hand off or channeling of the nicked DNA intermediate from nucleotide insertion step to following ligation step during BER pathway. Using this protocol, one is also able to measure ligation failure, or abortive ligation, after pol β oxidized nucleotide (8-oxo-dGTP) insertion. This is achieved by quantification of the addition of an adenylate (AMP) group at 5’-end of the BER intermediate. This protocol can also be used to measure nucleotide insertion coupled with ligation using other DNA polymerases and DNA ligases.
Materials and Reagents
- Eppendorf tubes (1.5 ml)
- Pipette tips (10 μl, 100 μl, 1,000 μl)
- Deoxyguanosine triphosphate (dGTP) (New England Biolabs, catalog number: N0447S )
- 8-oxo-2’-deoxyguanosine-5’-Triphosphate (8-oxo-dGTP) (Jena Bioscience, catalog number: NU-1117L )
- DNA substrate: The DNA substrate includes a fluorescent tag at both the 5’- and 3’-ends of one of the gap-containing strand in the double-stranded DNA with a single nucleotide gap opposite template base Cytosine (Çağlayan et al., 2017)
Note: The sequence information for the DNA substrate is presented in the Notes section of the protocol. - Tris-HCl (pH 7.5)
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P9333 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- Adenosine 5’-Triphosphate (ATP) (New England Biolabs, catalog number: P0756S )
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632 )
- Bovine serum albumin (BSA) (New England Biolabs, catalog number: B9000S )
- Glycerol (Sigma-Aldrich, catalog number: G9012 )
- HEPES (pH 7.5)
- Sodium chloride (NaCl)
- EDTA (Sigma-Aldrich, catalog number: 93283 )
- Purified enzymes: Recombinant human DNA polymerase β and DNA ligase I (see Recipes and Figure 2)
- Formamide (Sigma-Aldrich, catalog number: F9037 )
- Bromophenol blue (Sigma-Aldrich, catalog number: B0126 )
- Xylene cyanol (Sigma-Aldrich, catalog number: X4126 )
- Trizma-base (Sigma-Aldrich, catalog number: T4661 )
- Boric acid (Promega, catalog number: H5003 )
- Urea (National Diagnostic, catalog number: EC-605 )
- Ammonium persulfate (APS) (Sigma-Aldrich, catalog number: A3678-25G )
- Tetramethylethylenediamine (TEMED) (Sigma-Aldrich, catalog number: T9281-25ML )
- Sterile water
- AccuGel (40%) 19:1 Acrylamide to Bisacrylamide Stabilized Solution (National Diagnostic, catalog number: EC-850 )
- Full-range rainbow protein ladder (AmershamTM ECLTM RainbowTM Marker) (GE Healthcare, catalog number: RPN800E )
- 1x reaction buffer (see Recipes)
- 1x protein storage and dilution buffer (see Recipes)
- Enzyme mixture (see Recipes)
- Gel-loading dye (see Recipes)
- 10x TBE solution (see Recipes)
- Denaturing PAGE solution (15%) (see Recipes)
- EDTA buffer (pH 8.0) (see Recipes)
Equipment
- Pipettes (P2, P10, P20, P100, P200)
- Table-top heat block (Digital Dry Block Heater, VWR)
- Polyacrylamide gel electrophoresis (PAGE) apparatus (Biometra, model: Model S2 )
- Typhoon Phosphor Imager (GE Healthcare, model: Typhoon FLA 9500 )
Procedure
- Prepare 10 μl of reaction mixture (final volume) including 1 μl of 10x reaction buffer (1x final concentration), 2 μl of 1 mM dGTP or 8-oxo-dGTP (200 μM final concentration), and 1 μl of 2.5 μM DNA substrate (250 nM final concentration). Each reaction tube contains 10 µl of the reaction mixture needed for each time point. (see Recipes)
- Preincubate the enzyme mixture including 1 μl of 1 μM pol β and DNA ligase I (100 nM final concentration) at 37 °C for 3 min. (see Recipes)
- Start the reaction by addition of the enzyme mixture, prepared as above, to the reaction mixture, and pipette up and down to mix the reaction components.
- Incubate the reaction mixture at 37 °C for time points of 0.5, 1, 2, 3, 4, 5, 6, 8, 10, 12, and 15 min.
- Quench the reactions with 0.3 mM EDTA. (see Recipes)
- Mix the reaction products with an equal volume of gel loading dye.
- Heat the reaction samples at 95 °C for 3 min before loading onto the gel, and load 5 μl of reaction sample.
- Separate the reaction products in a 15% polyacrylamide gel containing 8 M urea at 30 mA for ~2 h.
- Scan the gel on a Typhoon Phosphor Imager as described (Çağlayan et al., 2014; 2015 and 2017).
Data analysis
A sample gel image that shows the products of pol β nucleotide insertion, DNA ligation by DNA ligase, and ligation failure is presented below (Figure 1). The original results were published in Çağlayan et al. (2017).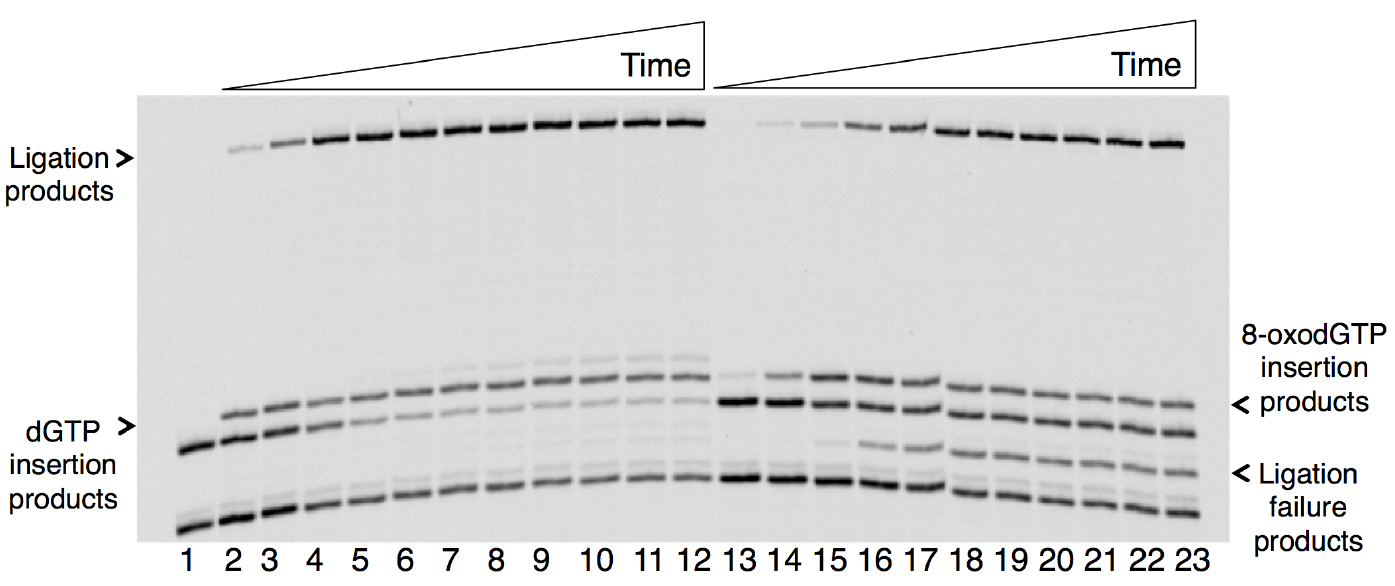
Figure 1. Time course-dependent changes in the products of nucleotide insertion and DNA ligation in a coupled BER assay in vitro. Lane 1 is a minus enzyme control, lanes 2-12 are the reaction products of dGTP insertion and ligation, and lanes 13-23 are the reaction products of 8-oxo-dGTP insertion, ligation and ligation failure. The reaction time points corresponding to lanes 2-12 and 13-23 are 0.5, 1, 2, 3, 4, 5, 6, 8, 10, 12, and 15 min. The sizes of DNA substrate for insertion and ligation are 18- and 16-mer, respectively. The sizes of the reaction products for dGTP or 8-oxo-dGTP insertion and ligation are 19- and 35-mer, respectively. The size of the abortive ligation product is 16-mer plus an adenylate (AMP) group (~17-mer).
Representative gel images (Figure 2) illustrate the purity of the human recombinant enzymes, DNA polymerase β (38 kDa) and DNA ligase I (101 kDa). The procedure for the purification of these enzymes was published previously (Beard and Wilson, 1995 and Chen Representative gel images (Figure 2) illustrate the purity of the human recombinant enzymes, DNA polymerase β (38 kDa) and DNA ligase I (101 kDa). The procedure for the purification of these enzymes was published previously (Beard and Wilson, 1995 and Chen et al., 2006). 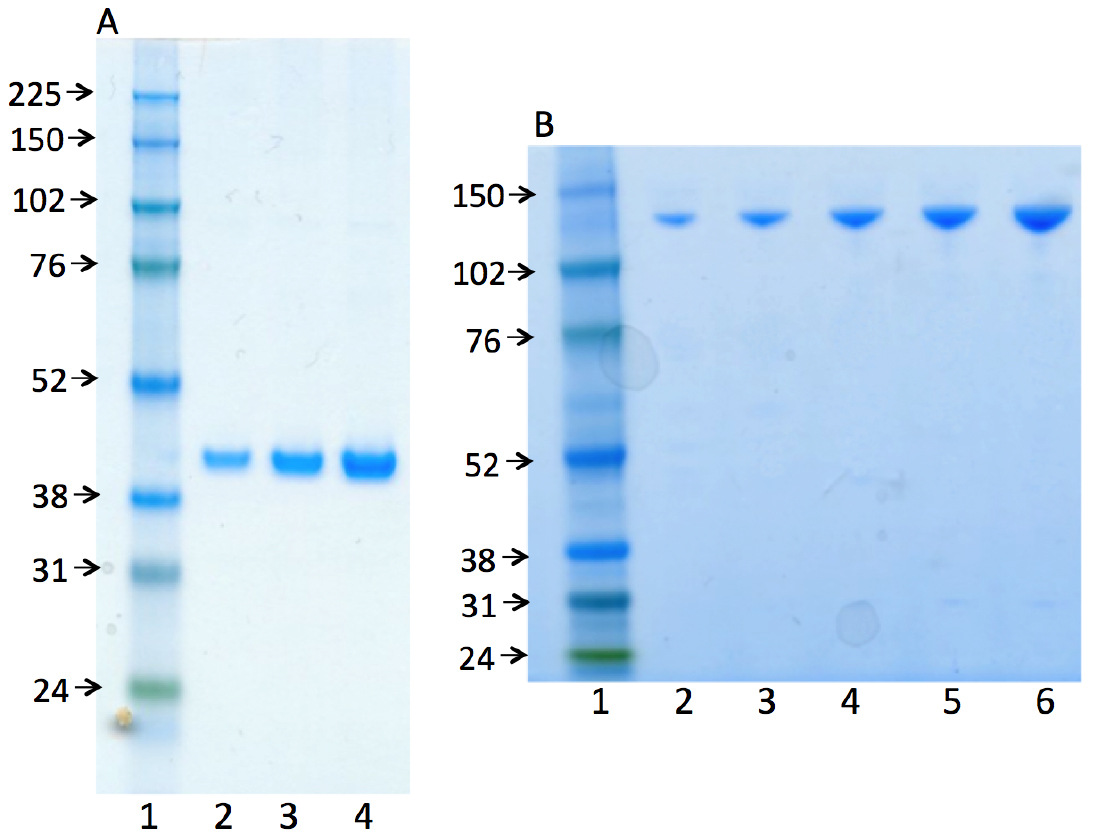
Figure 2. Photographs of SDS-PAGE analyses for the purified enzymes used, human recombinant DNA polymerase β (A) and human recombinant DNA ligase I (B). In both panels, lane 1 represents the marker protein ladder. Lanes 2-4 (in panel A) and lanes 2-6 (in panel B) are serial dilutions of pol β and ligase I, respectively.
Notes
- After preincubation of the enzyme mixture including pol β and DNA ligase I, the reaction should be immediately started by mixing this enzyme mixture into the reaction tube including 1x reaction buffer, dNTP, and DNA substrate. In addition, the enzymatic activities are determined with the substrate in excess over enzyme, under steady-state conditions.
- The oligonucleotides (Integrated DNA Technologies, IDT) below are used to construct a double-stranded DNA substrate, including a single-nucleotide gap in one strand (Çağlayan et al., 2017). FAM indicates a fluorescent label at 5’- or 3’-end of the upstream or downstream oligonucleotide.
Upstream oligonucleotide: FAM-5’-AACATGGGCGGCATGAAAT
Downstream oligonucleotide: AATGCCCATCCTCACCA-3’-FAM
Template oligonucleotide: 3’-TTGTACCCGCCGTACTTTACTTACGGGTAGGAGTGGT-5’
Recipes
- 1x reaction buffer
50 mM Tris-HCl (pH 7.5)
100 mM KCl
10 mM MgCl2
1 mM ATP
1 mM DTT
100 μg/ml BSA
10% glycerol - 1x protein storage and dilution buffer
50 mM HEPES (pH 7.5)
100 mM NaCl
1 mM EDTA
0.1% BSA
20% glycerol - Enzyme mixture
1 mg/ml DNA polymerase β
0.5 mg/ml DNA ligase I
Prepare 1 μM dilutions for each enzyme in the dilution buffer above - Gel-loading dye
95% formamide
20 mM EDTA
0.02% bromophenol blue
0.02% xylene cyanol - 10x TBE solution
108 g Tris-base
55 g boric acid
40 ml 0.5 M EDTA
dH2O up to 1 L - Denaturing PAGE solution (15%)
40 g urea
8 ml 10% TBE solution
10 ml dH2O
250 μl 10% APS
35 μl TEMED - EDTA buffer (pH 8.0)
10 mM Tris-HCl
1 mM disodium EDTA
Acknowledgments
This work was supported by the Intramural Research Program of the US National Institutes of Health, National Institute of Environmental Health Sciences (grants Z01 ES050158 and ES050159).
References
- Beard, W. A. and Wilson, S. H. (1995). Purification and domain-mapping of mammalian DNA polymerase β. Methods Enzymol 262: 98-107.
- Çağlayan, M., Batra, V. K., Sassa, A., Prasad, R. and Wilson, S. H. (2014). Role of polymerase beta in complementing aprataxin deficiency during abasic-site base excision repair. Nat Struct Mol Biol 21(5): 497-499.
- Çağlayan, M., Horton, J. K., Dai, D. P., Stefanick, D. F. and Wilson, S. H. (2017). Oxidized nucleotide insertion by pol β confounds ligation during base excision repair. Nat Commun 8: 14045.
- Çağlayan, M., Horton, J. K., Prasad, R. and Wilson, S. H. (2015). Complementation of aprataxin deficiency by base excision repair enzymes. Nucleic Acids Res 43(4): 2271-2281.
- Chen, X., Pascal, J., Vijayakumar, S., Wilson, G. M., Ellenberger, T. and Tomkinson, A. E. (2006). Human DNA ligases I, III, and IV-purification and new specific assays for these enzymes. Methods Enzymol 409: 39-52.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Çağlayan, M. and Wilson, S. H. (2017). In vitro Assay to Measure DNA Polymerase β Nucleotide Insertion Coupled with the DNA Ligation Reaction during Base Excision Repair. Bio-protocol 7(12): e2341. DOI: 10.21769/BioProtoc.2341.
Category
Molecular Biology > DNA > DNA damage and repair
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










