- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protein Isolation from Plasma Membrane, Digestion and Processing for Strong Cation Exchange Fractionation
Published: Vol 7, Iss 10, May 20, 2017 DOI: 10.21769/BioProtoc.2298 Views: 11579
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2869 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1989 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1772 Views
Abstract
Plasma membrane (PM) proteins play crucial roles in diverse biological processes. But their low abundance, alkalinity and hydrophobicity make their isolation a difficult task. This protocol describes an efficient method for PM proteins isolation, digestion and fractionation so that they can be well prepared for mass spectrometry analysis.
Keywords: Plasma membrane proteinsBackground
Plasma membrane (PM) proteins participate in diverse biological processes including signal transduction, ion transport and membrane trafficking, and are the first responders in cell-environment communication. They have a complicated composition varying from species, cell types and developmental stages (Alexandersson et al., 2008). Revealing their components and the expression features comprehensively with mass spectrometry (MS) is of great importance for developmental biology. However, their hydrophobic nature and the low abundance are a big challenge for the proteomic analysis (Wu and Yates, 2003). Additives like normal surfactants, organic solvents and urea are often used to improve PM proteins’ solubility, but they will reduce the proteases’ activities and create ion suppression during MS analysis (Zhang, 2015). RapiGest SF is a novel acid-labile anionic surfactant, which is structurally and functionally similar to SDS but does not inhibit the common endopeptidases activities at low concentration (0.1% w/v). Thus RapiGest SF used in solubilizing PM proteins can not only facilitate their digestion by exposing cleavage sites but is also easily quenched by strong acid and removed through centrifugation so that the surfactant does not affect the MS identification (Yu et al., 2003). Peptides yield by the RapiGest SF-assisted digestion can directly undergo the strong cation exchange (SCX) fractionation (Yang and Wang, 2017), so that the low abundance peptides can be detected by the MS.
Materials and Reagents
- 1.5 ml tubes (Corning, Axygen®, catalog number: MCT-150-C )
- ZipTip® pipette tip (Merck Millipore, catalog number: ZTC18S096 )
- PierceTM Spin columns, screw cap (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 69705 )
- C18 (FUJIGEL HANBAI, catalog number: MB 100 - 40/75 )
- Tris (2-carboxyethyl) phosphine hydrochloride solution, 0.5 M, pH 7.0 (TCEP) (Sigma-Aldrich, catalog number: 646547 )
- Formic acid (Sigma-Aldrich, catalog number: 94318 )
- Acetonitrile (Sigma-Aldrich, catalog number: 34851 )
- RapiGest SF surfactant (WATERS, catalog number: 186001861 )
- Iodoacetamide (Sigma-Aldrich, catalog number: I1149 )
- Trypsin (Roche Diagnostics, catalog number: 11418025001 )
- Potassium phosphate dibasic trihydrate (K2HPO4·3H2O)
- Potassium phosphate monobasic (KH2PO4)
- Phosphoric acid (H3PO4)
- Ammonium chloride (Sigma-Aldrich, catalog number: A9434 )
- Hydrochloric acid (HCl)
- 0.5 M iodoacetamide stock (see Recipes)
- 0.1 μg/μl trypsin stock (see Recipes)
- 50 mM potassium phosphate buffer, pH 7.8 (see Recipes)
- 100 ml solvent A (5 mM NH4Cl, 25% [v/v] acetonitrile, pH 3.0) (see Recipes)
- 100 ml solvent B (500 mM NH4Cl, 25% [v/v] acetonitrile, pH 3.0) (see Recipes)
Equipment
- 2 μl, 10 μl, 100 μl, 1,000 μl Pipetman (Gilson, France)
- Optimal MAX-XP ultracentrifuge (Beckman Coulter, model: Optimal MAX-XP )
- AKTA purifier-10 (GE Healthcare, model: AKTApurifier 10 )
- ISS 110 SpeedVac System (Thermo Fisher Scientific, Thermo ScientificTM, model: ISS 110 )
- PolySULFOETHYL ATM columns, 2.1 x 200 mm, 5 μm particles, 300 Å poresize (PolyLC, catalog number: 202SE05 )
Software
- UNICORN 5.2 software (GE Healthcare, USA)
Procedure
- Plasma membrane protein preparation
- Suspend the plasma membrane (PM) pellets (isolated according to Han et al., 2010) with 100 μl 50 mM potassium phosphate buffer, pH 7.8.
Note: The recommended PM protein concentration is 1 μg/μl. This protocol set 100 μg PM protein as an example, so use 100 μl buffer to suspend the pellets. - Add 25 μl 1% (w/v) RapiGest SF stock to the working concentration of 0.2% (w/v), mix well and then boil the mixture at ~100 °C for 5 min to dissolve proteins from PM.
Note: RapiGest SF is an acid labile denaturant (Figure 1A) which could solubilize and unfold proteins to make them more amenable for cleavage (Figure 1B). The recommended RapiGest SF working concentration is 0.1% (w/v). For PM proteins increase the incubation time (at 37 °C, up to 1 h is safe; at 100 °C, 5 min is enough) or use higher RapiGest SF concentration (up to 0.5% [w/v] is safe) could make the following digestion more efficient.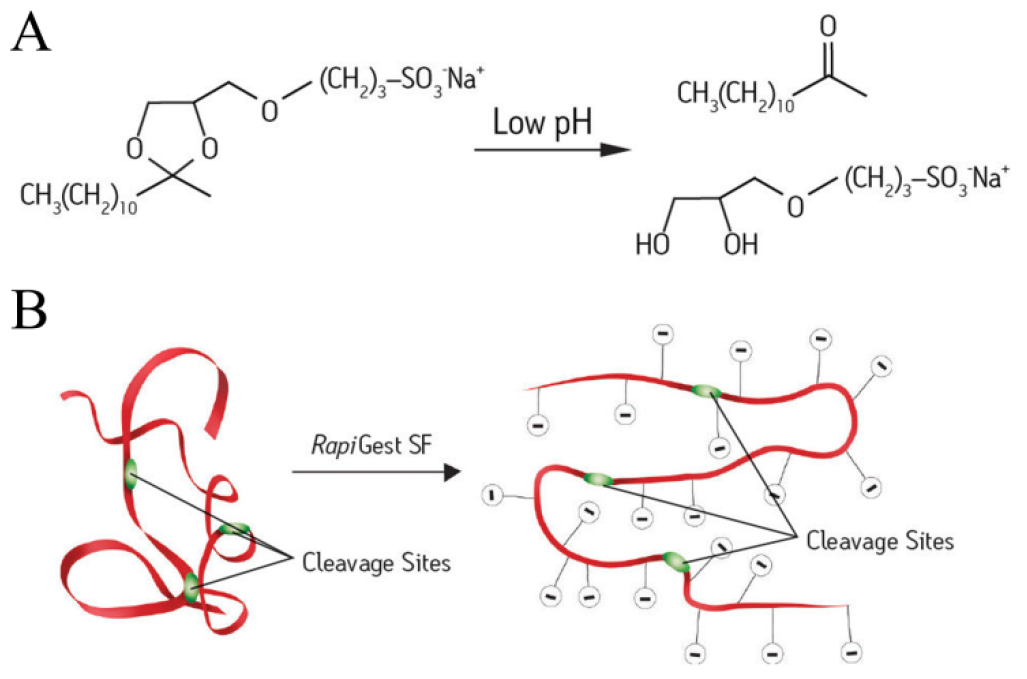
Figure 1. Working principle of RapiGest SF surfactant. A. RapiGest SF can be easily broken down by acidification and then removed by centrifugation. B. RapiGest SF is a mild denaturant which making proteins more amenable for cleavage without disrupting the endoproteases activity. Pictures were downloaded from the official website of Waters: (http://www.waters.com/waters/en_US/RapiGestSFSurfactant/nav.htm?locale=/&cid=1000941) - Cool down the mixture to room temperature (RT) for the following digestion.
- Suspend the plasma membrane (PM) pellets (isolated according to Han et al., 2010) with 100 μl 50 mM potassium phosphate buffer, pH 7.8.
- Plasma membrane protein digestion
- Reduction: add 2.5 μl 0.5 M TCEP, pH 7.0 to the protein mixture to a working concentration of 10 mM, mix them gently and then incubate at 56 °C for 1 h.
- Alkylation: cool down the mixture to RT, and add 14 μl 0.5 M iodoacetamide stock to a working concentration of 50 mM, mix them gently and then incubate at RT, dark, for 45 min.
- Digestion: add 20 μl 0.1 μg/μl trypsin stock to the protein mixture to a working concentration of 1:50 (w/w), mix them gently and then incubate at 37 °C for 12 h.
- Acidification: add 0.5 μl formic acid to the mixture to a working concentration of 0.2-0.5% (v/v) (pH < 2), incubate at 37 °C for 30-45 min, then centrifuge at 4 °C, 20,000 x g for 20 min to remove RapiGest SF. Discharge the pellets, transfer the supernatant to a new tube.
- Desalting: use C18 filled spin column to concentrate and purify the peptides mixture.
Note: If the peptides mixture is less than 5.0 μg, please use ZipTip® pipette tip to do the desalting. - Put the purified peptides mixture into SpeedVac system to lyophilize for the following fractionation use.
- Reduction: add 2.5 μl 0.5 M TCEP, pH 7.0 to the protein mixture to a working concentration of 10 mM, mix them gently and then incubate at 56 °C for 1 h.
- Peptides mixture fractionation
The following protocol was modified from Zhu et al. (2009). It performs on the AKTA purifier-10 system with the strong cation exchange (SCX) PolySULFOETHYL ATM column.- Resuspend the lyophilized peptides mixture in solvent A.
- Run solvent B for 20 min, then solvent A for 30 min through the AKTA purifier-10 system to balance the PolySULFOETHYL ATM column.
- Load the suspended peptides and run the following gradient at a flow rate of 200 μl/min, collect the products in every 2 min.
0-5 min: 100% solvent A
5-95 min: 0-60% solvent B
95-115 min: 60%-100% solvent B
115-145 min: 100% solvent B
145-155 min: 100% solvent A - Run ddH2O for 20 min, then 100% acetonitrile for 10 min to store the column.
- Pool the products according to the 214 nm UV absorption peak (Figure 2).
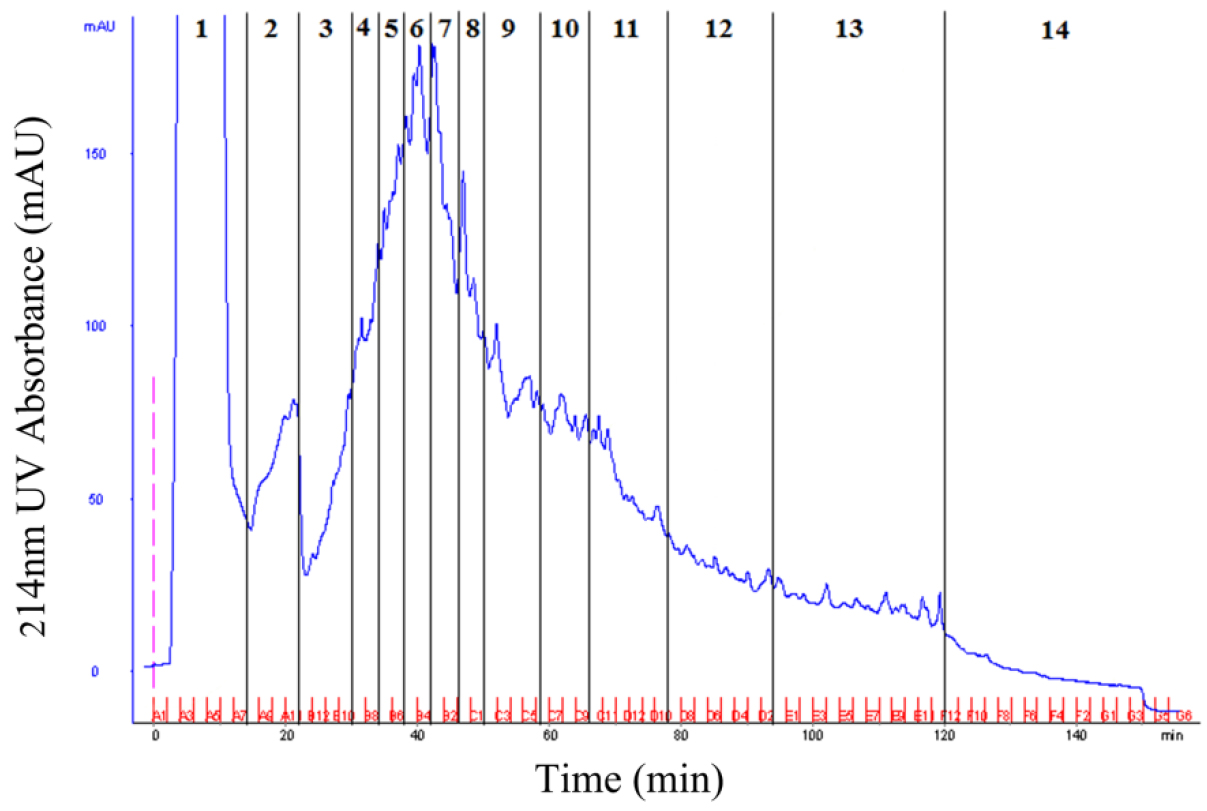
Figure 2. SCX fractionation of peptides mixture from rice mature pollen grain. According to the 214 nm UV absorption peak, the peptides mixture can be pooled into 14 fractions for LC-MS/MS analysis. - Use C18 spin column or ZipTip® pipette tip to purify the combined products.
- Lyophilize the purified products for mass spectrometry identification.
- Resuspend the lyophilized peptides mixture in solvent A.
Data analysis
The SCX separated peptides were monitored by their 214 nm UV absorption peak through the UNICORN 5.2 software (GE Healthcare, USA).
Notes
After SCX fractionation, the repooled peptides contains a high concentration of salts. Don’t forget to do the desalting procedure through C18 spin column or ZipTip® pipette tip, or the following mass spectrometry identification will be affected.
Recipes
- 1% (w/v) RapiGest SF stock
Dissolve 1 mg of lyophilized RapiGest SF powder in 100 μl ddH2O, store at -20 °C - 0.5 M iodoacetamide stock
Dissolve 46 mg of iodoacetamide powder in 500 μl ddH2O, use immediately - 0.1 μg/μl trypsin stock
Dissolve 25 μg of trypsin powder in 250 μl ddH2O, store at -20 °C - 200 ml 50 mM potassium phosphate buffer, pH 7.8
2.072 g K2HPO4·3H2O
0.125 g KH2PO4
Fill up to 200 ml with ddH2O
Adjust pH to 7.8 with H3PO4 - 100 ml solvent A (5 mM NH4Cl, 25% [v/v] acetonitrile, pH 3.0)
0.027 g NH4Cl
25 ml acetonitrile
Fill up to 100 ml with ddH2O
Adjust pH to 3.0 with HCl - 100 ml solvent B (500 mM NH4Cl, 25% [v/v] acetonitrile, pH 3.0)
2.675 g NH4Cl
25 ml acetonitrile
Fill up to 100 ml with ddH2O
Adjust pH to 3.0 with HCl
Acknowledgments
This protocol was mainly modified from Han et al. (2010) and Yang and Wang (2017). This work was supported by the Chinese Ministry of Science and Technology (grant No. 2013CB945101) and the China Postdoctoral Science Foundation (grant No. 2016M591284).
References
- Alexandersson, E., Gustavsson, N., Bernfur, K., Karlsson, A., Kjellbom, P. and Larsson, C. (2008). Purification and proteomic analysis of plant plasma membranes. Methods Mol Biol 432: 161-173.
- Han, B., Chen, S., Dai, S., Yang, N. and Wang, T. (2010). Isobaric tags for relative and absolute quantification- based comparative proteomics reveals the features of plasma membrane-associated proteomes of pollen grains and pollen tubes from Lilium davidii. J Integr Plant Biol 52(12): 1043-1058.
- Wu, C. C. and Yates, J. R., 3rd (2003). The application of mass spectrometry to membrane proteomics. Nat Biotechnol 21(3): 262-267.
- Yang, N. and Wang, T. (2017). Comparative proteomic analysis reveals a dynamic pollen plasma membrane protein map and the membrane landscape of receptor-like kinases and transporters important for pollen tube growth and interaction with pistils in rice. BMC Plant Biol 17(1): 2.
- Yu, Y. Q., Gilar, M., Lee, P. J., Bouvier, E. S. and Gebler, J. C. (2003). Enzyme-friendly, mass spectrometry-compatible surfactant for in-solution enzymatic digestion of proteins. Anal Chem 75(21): 6023-6028.
- Zhang, X. (2015). Less is more: Membrane protein digestion beyond urea-trypsin solution for next-level proteomics. Mol Cell Proteomics 14(9): 2441-2453.
- Zhu, M., Dai, S., McClung, S., Yan, X. and Chen, S. (2009). Functional differentiation of Brassica napus guard cells and mesophyll cells revealed by comparative proteomics. Mol Cell Proteomics 8(4): 752-766.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yang, N., Han, B. and Wang, T. (2017). Protein Isolation from Plasma Membrane, Digestion and Processing for Strong Cation Exchange Fractionation. Bio-protocol 7(10): e2298. DOI: 10.21769/BioProtoc.2298.
Category
Plant Science > Plant biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












