- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Mimicking Angiogenesis in vitro: Three-dimensional Co-culture of Vascular Endothelial Cells and Perivascular Cells in Collagen Type I Gels
Published: Vol 7, Iss 8, Apr 20, 2017 DOI: 10.21769/BioProtoc.2247 Views: 18965
Reviewed by: Joseph C. ChenAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Differentiation, Maintenance, and Contraction Profiling of Human Induced Pluripotent Stem Cell–Derived Cardiomyocytes
Matthijs Snelders [...] Jeroen Essers
Mar 5, 2025 3937 Views

Isolation and Culture of Ferret Airway Stem Cells
Ziying Yan [...] Feng Yuan
Jul 20, 2025 2413 Views

Optimization of Adipogenic Differentiation Protocol for Murine and Human Cell Culture Models
Junwan Fan [...] Wenyan He
Jan 20, 2026 230 Views
Abstract
Angiogenesis defines the process of formation of new vascular structures form existing blood vessels, involved during development, repair processes like wound healing but also linked to pathological changes. During angiogenic processes, endothelial cells build a vascular network and recruit perivascular cells to form mature, stable vessels. Endothelial cells and perivascular cells secret and assemble a vascular basement membrane and interact via close cell-cell contacts. To mimic these processes in vitro we have developed a versatile three-dimensional culture system where perivascular cells (PVC) are co-cultured with human umbilical cord vascular endothelial cells (HUVEC) in a collagen type I gel. This co-culture system can be used to determine biochemical and cellular processes during neoangiogenic events with a wide range of analyses options.
Keywords: Endothelial cellsBackground
The coordinated interaction between endothelial and perivascular cells is essential to form a stable vascular network according to the local needs within a given tissue. Multiple molecular components contribute to the interactions but are still poorly understood. Various growth factors are needed to attract endothelial cells to sites of low oxygen concentrations and build new vessels that are then covered by perivascular cells. Both cell types interact to secrete a specialized extracellular matrix and stabilize the newly formed vessels. In the past multiple assays have been established to analyze vascular cell interaction and vessel-like network formation on two-dimensional matrigel substrates, but those are limited in providing information about initial steps of endothelial-perivascular cell interaction and vascular basement membrane formation in a three-dimensional microenvironment. In addition, well characterized perivascular cell suitable for culture experiments were missing.
We have previously isolated cells with perivascular characteristics, as they express pericyte-specific markers, produce and secrete extracellular matrix proteins and stimulate angiogenic processes in vivo (Brachvogel et al., 2005 and 2007). These cells were used to establish a co-culture system with human umbilical vein endothelial cells and study critical steps in neoangiogenesis upon interaction of the two cell types in a three-dimensional microenvironment (Pitzler et al., 2016; Zhou et al., 2016).
The co-cultures showed a superior activity to promote the formation endothelial tube formation and stabilization by perivascular cell recruitment and basement membrane formation. The culture system allows to monitor migration and interaction of vascular cells by time-lapse microscopy and to study the deposition of basement membrane protein by immunofluorescence analysis. To quantify and isolate endothelial and perivascular cells from co-cultures immunomagnetic or flow cytometry sorting approaches can be used and cell type-specific global changes in mRNA and miRNA expression can be analyzed by subjecting isolated RNA from the separated cell populations to microarray analysis or RNA sequencing. Moreover, the effects of anti- and pro-angiogenic substances on endothelial-perivascular cell interaction can be analyzed in vitro. Hence, the three-dimensional culture protocol allows to study cellular, biochemical and transcriptional events during neoangiogenesis and to characterize the pro- and anti-angiogenetic effects of molecules on endothelial perivascular cell interaction ex vivo.
Materials and Reagents
- 15 ml tubes (Greiner Bio One International, catalog number: 188271 )
- 100 mm TC-treated cell culture dish (Corning, Falcon®, catalog number: 353003 )
- 1.5 ml tubes (SARSTEDT, catalog number: 72.690.001 )
- 24-well-plate (Corning, Costar®, catalog number: 3524 )
- T75-flask (VWR, catalog number: 734-0050 )
- 48-well-plate (Corning, Costar®, catalog number: 3548 )
- 10 ml syringe (BD, catalog number: 309604 )
- Sterile syringe filter with a 0.22 µm pore size (EMD Millipore, catalog number: SLGP033RB )
- Disposable HSW FINE-JECT® needles (Henke Sass Wolf, catalog number: 4710004012 )
- Pipettes Gilson (10 µl, 20 µl, 100 µl, 1,000 µl, 5 ml)
- Macro pipet tips (VWR, catalog number: 89368-994 )
- Microscope cover glasses: squares (Fisher Scientific, catalog number: S175222 )
- Adhesion slides, Menzel Gläser, SuperFrost® Plus (VWR, catalog number: 631-9483 )
- HUVEC (human umbilical cord vascular endothelial cell) (Lonza, catalog number: CC-2519 or self-isolation of HUVECs according to Baudin et al., 2007)
- Isolated perivascular cells/pericytes (PVC) (see Note 1)
- AnxA5-LacZ mice (Anxa5tm1Epo) (Brachvogel et al., 2003)
- Collagenase type II (Worthington Biochemical, catalog number: LS004176 )
- DNase I recombinant (Roche Diagnostics, catalog number: 04536282001 )
- Fluorescein-di-(β-D-galactopyranoside) (FDG) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: F1179 )
- Propidium iodide (PI) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: P3566 )
- Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific, GibcoTM, catalog number: 31966021 )
- Fetal calf serum (FCS) (Biochrom, catalog number: S 0115 )
- Penicillin-streptomycin (P/S) (10,000 U/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- Gelatin from porcine skin (Sigma-Aldrich, catalog number: G2500 )
- Sodium bicarbonate (NaHCO3) (EMD Millipore, catalog number: 1.6329.1000 )
- Rat tail type I collagen solution (Corning, catalog number: 354236 )
- Sodium hydroxide (NaOH) (VWR, catalog number: 28244.295 )
- Phosphate-buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 18912014 )
- Trypsin/EDTA (Biochrom, catalog number: L 2153 )
- Endothelial growth medium 2 (EGM2) (PromoCell, catalog number: C-22111 )
- Recombinant VEGF-A/Vascular endothelial growth factor (Biomol, catalog number: 94900 )
- Recombinant PDGF/Platelet-derived growth factor-BB (Biomol, catalog number: 94968 )
- Ascorbic acid phosphate (Sigma-Aldrich, catalog number: A8960-5G )
- Methanol (VWR, catalog number: 20903.461 )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D8418-100ML )
- Tween-20 (Sigma-Aldrich, catalog number: P1379-500ML )
- Bovine serum albumin (BSA) (SERVA Electrophoresis, catalog number: 11930.03 )
- Anti-human CD31 (BD, BD Biosciences, catalog number: 555444 )
- Anti-NG2-proteoglycan (EMD Millipore, catalog number: AB5320 )
- Donkey-anti-rat Ig, Cy2 conjugated (Jackson ImmunoResearch, catalog number: 712-225-150 )
- Donkey, anti-rabbit Ig, Cy2 conjugated (Jackson ImmunoResearch, catalog number: 711-225-152 )
- Donkey, anti-rabbit Ig, Cy5 conjugated (Jackson ImmunoResearch, catalog number: 711-175-152 )
- Donkey, anti-goat Ig, Cy2 conjugated (Jackson ImmunoResearch, catalog number: 705-225-147 )
- Mounting solution
- CFSE (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: C1157 )
- SNARF-1 (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: S22801 )
- MCDB-131 medium (Thermo Fisher Scientific, GibcoTM, catalog number: 10372019 )
- Basic fibroblast growth factor (bFGF) (ReliaTech, catalog number: 300-003 )
- Epidermal growth factor (EGF) (Biomol, catalog number: 97052 )
- Insulin solution human (Sigma-Aldrich, catalog number: I9278 )
- Heparin
- Hydrocortisone
- 10x DMEM, 10x Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, catalog number: D2429-100ML )
- Sodium pyruvate (Thermo Fisher Scientific, GibcoTM, catalog number: 11360070 )
- Trizma® base (Sigma-Aldrich, catalog number: T1503 )
- Sodium chloride (NaCl) (EMD Millipore, catalog number: 1064041000 )
- Autoclaved distilled H2O
- Proliferation medium (see Recipes)
- PVC culture medium (see Recipes)
- HUVEC culture medium (see Recipes)
- 2x medium-mix (see Recipes)
- Collagen solution (see Recipes)
- Tris-buffered saline (TBS) (see Recipes)
Equipment
- Centrifuge (Eppendorf, model: 5415 R )
- Hemocytometer (LO-Laboroptik, model: Neubauer-Improved )
- CO2 cell incubator (Heraeus)
- Centrifuge Labofuge (Heraeus Sepatech, model: Labofuge GL )
- Autoclave
- SteREO Lumar.V12 (Zeiss, model: SteREO Lumar.V12 )
- Axiovert 40 CFL (Zeiss, model: Axiovert 40 CFL )
- Axioplan 2 (Zeiss, model: Axioplan 2 )
- Water bath (Köttermann)
- Vortex mixer (Scientific Industries)
- Inverted microscope Eclipse TE2000-U (Nikon Instruments, model: Eclipse TE2000-U )
- Fluorescence-activated cell sorting (MoFlow, Cytomation)
- Biosafety cabinet LaminAir HB 2448 (Heraeus, model: LaminAir HB 2448 )
Software
- ImageJ (Fiji)
Procedure
- Original protocol for pericyte isolation from meninges using AnxA5-LacZ mice
- Prepare approximately 200 ml of 5% FCS-PBS and cool down 100 ml of 5% FCS-PBS on ice.
- Carefully isolate the brain from 4-6 months old AnxA5-LacZ mice. Dissect the meninges under a stereomicroscope and transfer them into a 15 ml tube with 5% FCS-PBS on ice. Collect several meninges and then centrifuge the sample for five min at 400 x g at 4 °C.
- Aspirate supernatant and dissolve pellet in 1.5-3.0 ml of 0.4% collagenase type II. Incubate sample for 60 min at 37 °C slightly shaking. Subsequently, centrifuge digested tissue for five min at 400 x g at room temperature.
- Aspirate supernatant and resuspend in 1.5-3.0 ml of 2% trypsin/100 U DNAse I. Then, incubate digested tissue for 30 min at 37 °C slightly shaking. Next, centrifuge for five minutes at 400 x g at room temperature.
- Resuspend cell pellet in 5% FCS-PBS and filter cell suspension through a 100 μm cell strainer. After that, centrifuge cell suspension for 5 min at 400 x g at room temperature.
- Aspirate supernatant and resuspend cell pellet in 5% FCS-PBS. Determine cell number by using a hemocytometer, then dissolve 1 x 106 cells in 20 µl of 5% FCS-PBS and distribute 20 µl of the cell suspension into new 1.5 ml tubes. Add 20 µl of 2 mM fluorescein-di-(β-D-galactopyranoside) (FDG) to 20 µl cell suspension and incubate mix at 37 °C for 75 sec. Add 500 µl of ice-cold 5% FCS-PBS to the cells and incubate on ice in the dark for 3 h. Use a non-stained sample as control for cell sorting.
- Centrifuge cell suspension for 5 min at 400 x g at RT and resuspend cell pellet in 500 µl of 5% FCS-PBS. To exclude dead cells, add propidium iodide (final concentration 1 µg/ml)
- Sort stained cells by fluorescence-activated cell sorting (MoFlow, Cytomation) and collect propidium iodide negative and FDG positive cells. Next, plate 5 x 104 of PI negative/FDG positive cells per well in proliferation medium (see Recipes) on gelatin-coated 24-well-plate and store cells in a 5% CO2 humidity incubator at 37 °C. Change medium every second day and expanse isolated cells.
- After initial expansion, the cells are cultured in DMEM with 10% FCS and 1% P/S. Cells should be further characterized by flow cytometry. Cells are positive for Sca-1, CD140b and α-SMA and negative for PECAM, CD45, CD34, F4/F80 and c-kit. Isolated Anxa5-LacZ(+) PVC from mouse meninges retain their perivascular cell phenotype and can be cultured without undergoing senescence for multiple passages (Brachvogel et al., 2007).
Note: An established and well characterized PVC cell line with superior angiogenic activity can be send to individual researcher upon request to perform coculture experiments with endothelial cells (Zhou et al., 2016). This eliminates the need to isolate the cells from mouse brain meninges.
- Prepare approximately 200 ml of 5% FCS-PBS and cool down 100 ml of 5% FCS-PBS on ice.
- Co-culture of PVCs and HUVECs in collagen type I gels
- Dissolve 0.1 g of gelatin in 100 g of autoclaved distilled water (final concentration 0.1%) and autoclave the solution at 121 °C for 30 min. Then, coat 100 mm TC-treated cell culture dish with gelatin by adding 5 ml of 0.1% gelatin solution to the dish and distribute gelatin solution until the whole 100 mm TC-treated cell culture dish surface is fully covered. After at less two hours’ incubation at 37 °C, the residual gelatin solution is aspirated. Subsequently, dry dish at room temperature before use.
- HUVECs are cultured in EGM2 culture medium (see Recipes) on gelatin-coated plates in a 5% CO2 humidity incubator at 37 °C. Cells are cultured until subconfluent (~80%) and further passaged (1:3 dilution). HUVEC are only used in early passages (passage 3-6). PVCs are cultured in DMEM with 10% FCS and 1% P/S (see Recipes) on T75-flasks or on 10 cm dishes and expand to about 80% confluency (see Note 2 and Figure 1).
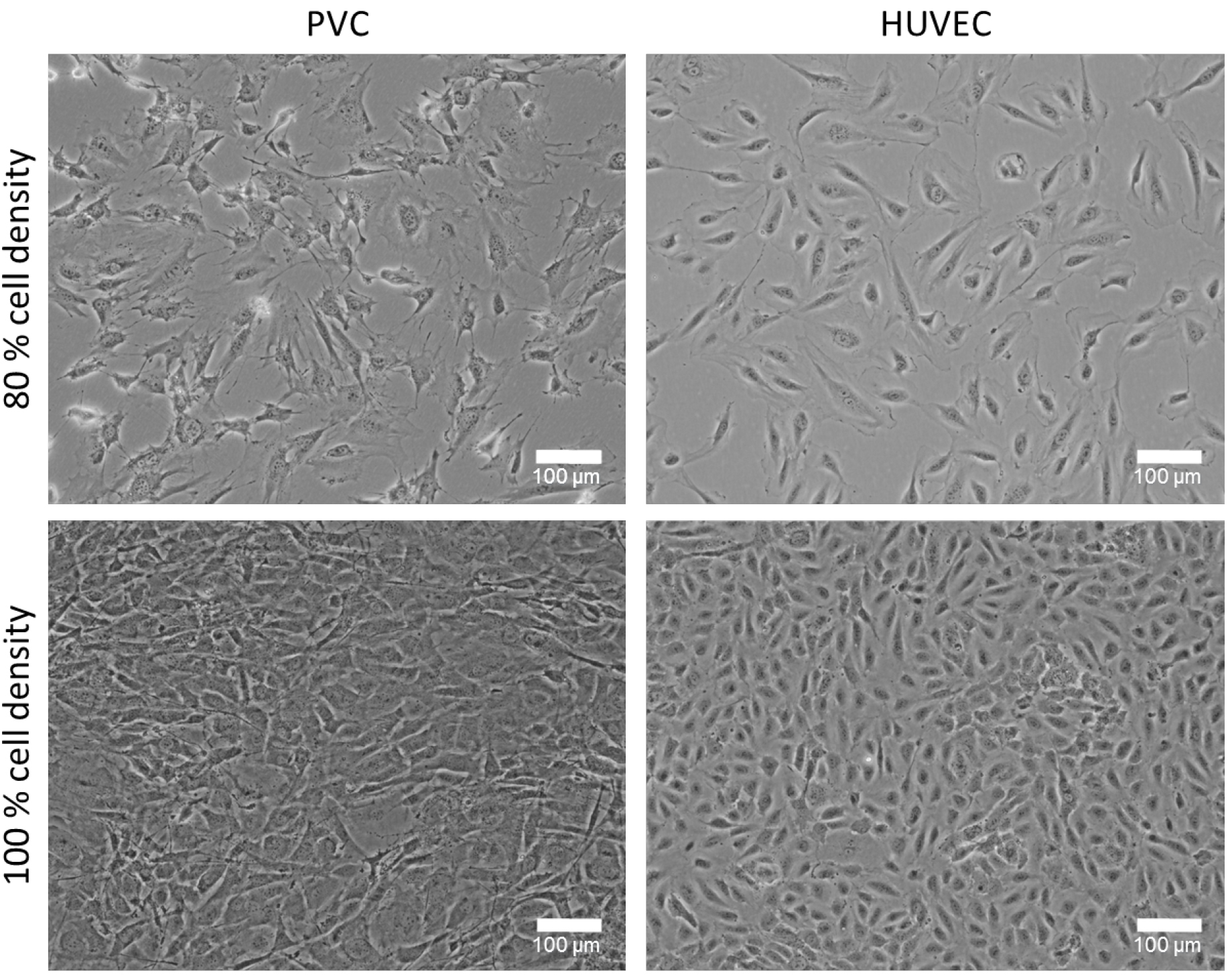
Figure 1. Cultured perivascular cells (PVC) and human umbilical cord vein endothelial cells (HUVEC) with different cell densities. Representative phase contrast images of 80% and 100% confluency (cell density) cultured PVCs and HUVECs. The cell density plays an important role for this co-culture experiment and can affect the tube formation. Bars = 100 µm. - Prepare 10 ml of 2x medium-mix (see Recipes). Therefore, dissolve 0.38 g of NaHCO3 in 10 ml of autoclaved distilled water (concentration 0.45 M) and after dissolving sterile filter the solution. Then, mix 2 ml of 10x DMEM, 200 µl of 100x sodium pyruvate and 1 ml of prepared 0.45 M NaHCO3 (final concentration 0.045 M) in 6.8 ml (final volume 10 ml) of autoclaved distilled H2O. Solution should be cool on ice until use.
- For 4 ml of collagen solution (see Recipes), mix 2 ml of 2x medium-mix, 800 µl FCS (final concentration 20%), 963 µl of 8.3 mg/ml rat tail type I collagen (see Note 3) (final concentration 2 mg/ml) and 23 µl of autoclaved distilled H2O on ice. Finally, neutralize collagen type I gel solution with 214 µl of 0.1 N NaOH on ice and adjust pH of solution to 7.4. Store the prepared collagen solution on ice until usage.
- Wash sub-confluent, cultured HUVECs and PVCs with 10 ml of PBS and detach cells with 1-2 ml trypsin/EDTA for 5 min at 37 °C. Add 9 ml of EGM2 culture medium to stop reaction and collect cells in a 15 ml tube.
- Centrifuge cells for 5 min at 400 x g at room temperature (RT). Aspirate supernatant and resuspend cell pellets in 5 ml of EGM2 culture medium. Add 10 µl of each cell suspension into a hemocytometer to determine cell concentration.
- For monoculture, transfer 6.25 x 105 HUVECs or 1.25 x 105 PVCs (see Note 4) in EGM2 medium in a new 15 ml tube. Mix 6.25 x 105 HUVECs and 1.25 x 105 PVCs for co-culture in 2 ml EGM2 medium in a new 15 ml tube (Figure 2). Alternatively, different endothelial cells or perivascular cells can be used, but the relative ratio of cells need to adapted and tested. Technical duplicates or triplicates are highly recommended. Centrifuge cell suspensions at 400 x g for 5 min at RT.
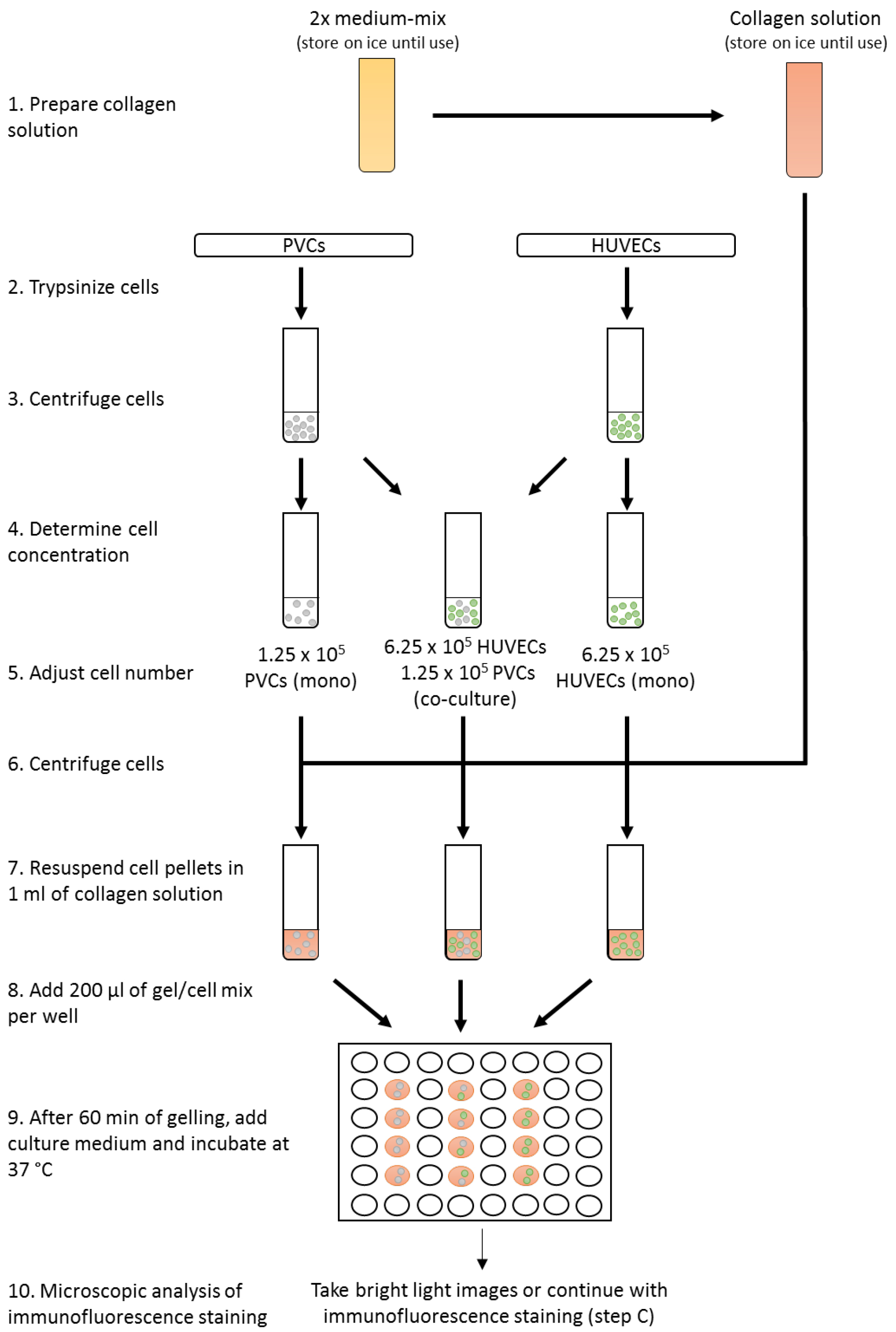
Figure 2. Workflow of the three-dimensional co-culture assay - Carefully remove all medium and resuspend cell pellets in 1 ml of collagen type I gel solution. Add 200 µl into each well of a 48-well plate and incubate for 60 min at 37 °C until solutions turn into gel. To increase homogeneity of gels, collagen type I gel solution with cells should be properly mixed and gel solution be uniformly disturbed in the well. Try to prevent uneven distribution of the solution by adding the gel solution directly in the center of the well and distribute by moving the 48-well plate carefully. To improve gelling, 48-well plates should be prewarmed in the incubator at 37 °C
- Check after 60 min whether a homogenous gel has formed. Otherwise incubate additional 30 min. After gelling, carefully add 400 µl EGM2 culture medium supplemented with 10 ng/ml VEGF, 10 ng/ml PDGF and 250 μg/ml ascorbic acid phosphate on top of each gel. Other growth factors or drugs may be added at this stage for different experimental conditions.
- Incubate the gels for 3-6 days (see Note 6) in a 5% CO2 humidity incubator and take images (Figure 3) or videos (3 images/h) every 24 h. We used 10 or 20x objectives to take images of networks (Zeiss SteREO Lumar.V12 or Zeiss Axiovert 40 CFL). Higher magnification can be used but the thickness of the gel can hinder the acquisition of images. EGM2 culture medium with supplements should be replaced every 48 h.
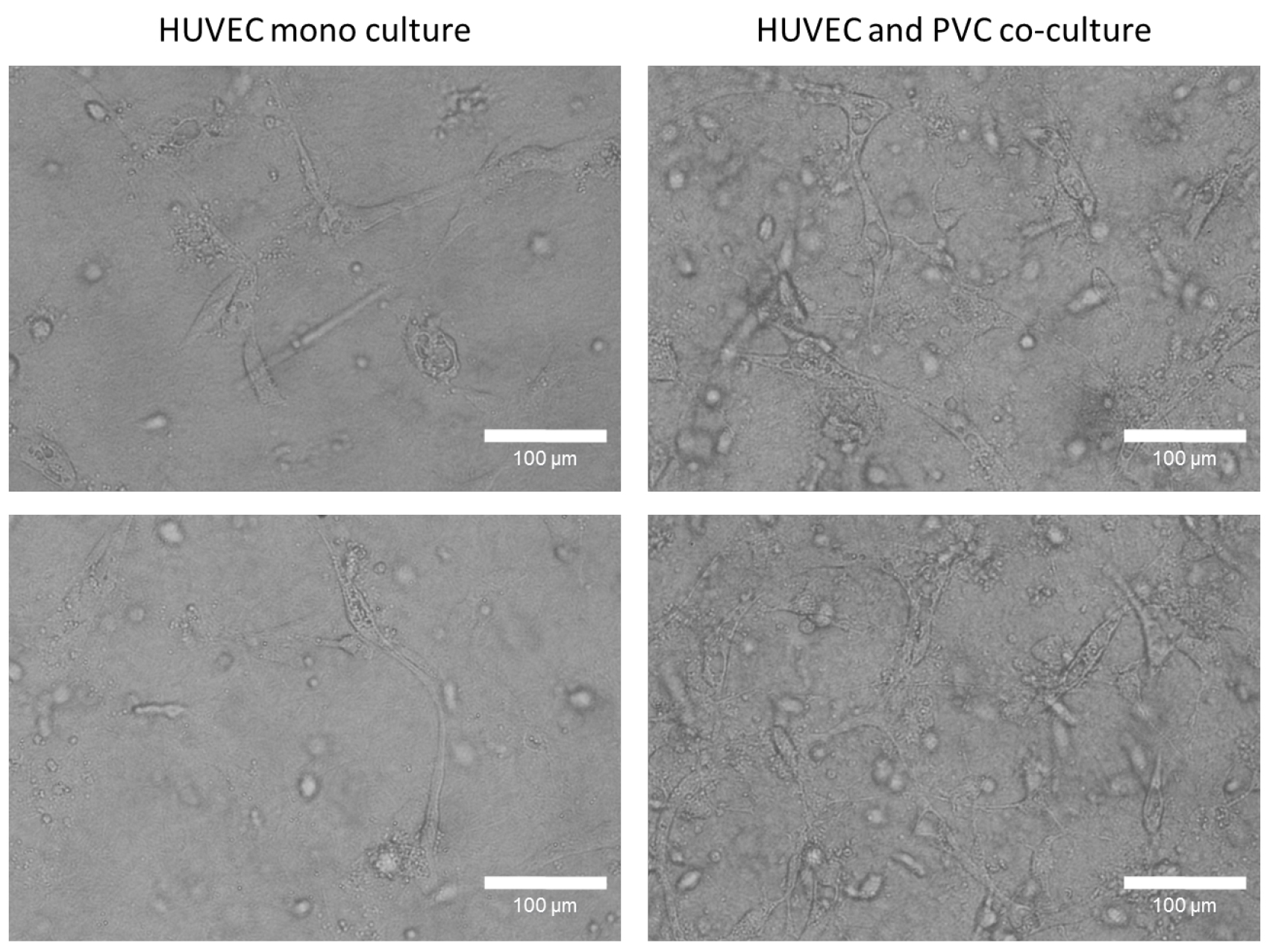
Figure 3. Tube formation studies of mono and co-culture HUVECs in a three-dimensional collagen type I gel. After 96 h, HUVECs form in present of perivascular cells a robust tube network in a collagen type I gel, whereas monocultured HUVECs few tubes. Representative phase contrast images were taken with 20x objective. Bars = 100 µm.
- Dissolve 0.1 g of gelatin in 100 g of autoclaved distilled water (final concentration 0.1%) and autoclave the solution at 121 °C for 30 min. Then, coat 100 mm TC-treated cell culture dish with gelatin by adding 5 ml of 0.1% gelatin solution to the dish and distribute gelatin solution until the whole 100 mm TC-treated cell culture dish surface is fully covered. After at less two hours’ incubation at 37 °C, the residual gelatin solution is aspirated. Subsequently, dry dish at room temperature before use.
- Immunohistochemical detection of extracellular matrix and cellular proteins in 3D cultures
- Wash gels twice with 600 µl of PBS (see Note 6).
- Fix gels with 400 µl of 80% methanol/20% DMSO (Dent’s Fixative) for 30 min at RT.
- Rehydrate gels with 600 µl of 50% methanol/PBS for 1 h at RT. Remove solution and add 600 µl of 20% methanol/PBS. Next, wash gel in 600 µl of PBS-T (PBS, 0.1% Tween-20) for 1 h at RT.
- Aspirate solution and add 600 µl of 10% FCS, 5% BSA in PBS for 2-4 h at RT to block non-specific antibody-binding.
- Carefully remove blocking solution and incubate gels with 150-300 µl of primary antibody diluted in blocking solution (10% FCS, 5% BSA in PBS), overnight at 4 °C lightly shaking on a rocking plate. Antibody dilutions should be tested empirically before.
- Wash gels with 600 µl of TBS-T (0.1% Tween in TBS) for an hour. Repeat washing steps six times.
- Incubate samples with 150-300 µl of fluorescent chromophore-conjugated, secondary antibodies in blocking solution for 2-16 h in the dark at RT.
- Wash gels with 600 µl of TBS-T (0.1% Tween in TBS) for an hour. Then, repeat washing step six times.
- Carefully remove gels from the wall of the well using a 27 G needle. Use a 5 ml macro pipet tips to transfer gels on slides. Add 50 µl of mounting solution on top of the gels and carefully mount samples with a glass coverslip.
- Images are taken with Zeiss Axioplan 2 by using 5x and 63x objectives and adequate filter sets to get overview and close-up images, respectively. To visualize HUVECs in three-dimensional cultures gels can be immunostained with CD31 (Figures 4A and 4B) and PVCs can be immunostained with NG2-PG (Figure 4B) or α-SMA specific antibodies. The overview images of CD31+ HUVECs are used for tube formation analysis.
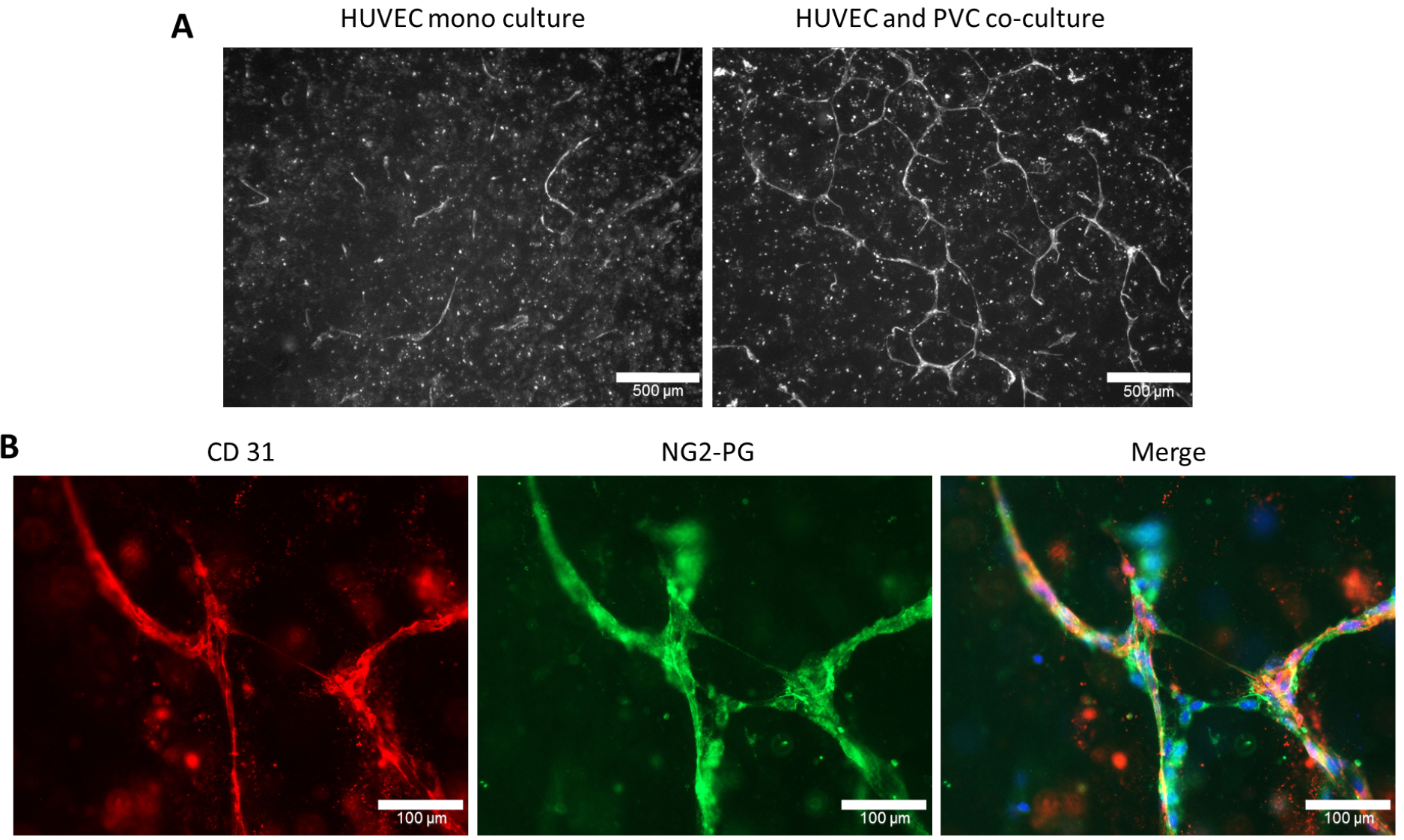
Figure 4. Endothelial tube formation of human umbilical cord vein endothelial cells (HUVEC) was induced upon co-coculturing with PVC. A. HUVECs are cultured either alone (monoculture) or in co-culture with PVC in a three-dimensional collagen I gel. After 6 days in culture, the three-dimensional collagen I gels were stained with human CD31 as described in Procedure C. Scale bars = 500 µM. B. Immunostainings of co-cultured HUVECs and PVCs using CD31 and NG2-proteoglycan (NG2-PG) specific antibodies. Individual or as merged images with nuclear staining (DAPI) are shown. Scale bars = 100 µm.
- Wash gels twice with 600 µl of PBS (see Note 6).
- Cell labelling with fluorescent dyes (CFSE, SNARF-1) for live cell tracking
- Dissolve CFSE or SNARF-1 in DMSO to a final concentration of 5 mM at RT in the dark. After using fluorescent dyes, solution can be aliquoted and stored at -20 °C. For subsequent use of fluorescent dyes, thaw CFSE and SNARF-1 dye at RT in the dark. Freshly prepare 100 ml of 3% FCS-PBS solution. Store 25 ml at RT and cool the remaining 75 ml on ice for at least 30 min.
- Detach HUVECs and PVCs as described (see step A3) and collect cells in 15 ml tubes. Determine cell number using a hemocytometer, then centrifuge cells for 5 min at 400 x g at RT.
- Resuspend cell pellets in an appropriate amount of 3% FCS-PBS to a final concentration of 1 x 107 cells per 1 ml of 3% FCS-PBS. Do not use less than 50 µl of 3% FCS-PBS solution.
- Prior to use, prepare 0.5 mM solutions of CFSE and SNARF-1 using PBS. Then, further dilute CFSE or SNARF-1 solutions (0.5 mM) using cell suspensions to a final concentration of 0.01 mM. Typically, 2 µl of 0.5 mM CFSE are added to 100 µl cell suspension. All following steps are performed in the dark avoiding light exposure.
- Mix HUVECs and PVCs for 10 sec at maximum speed on a vortex mixer. Then, incubate cell suspensions for 10 min in a water bath, pre-heated to 37 °C, without shaking.
- Add 5 ml of ice cold 3% FCS-PBS to stop the reaction and centrifuge cells for 5 min at 400 x g at 4 °C. Aspirate supernatant and resuspend pellets in 5 ml of ice cold 3% FCS-PBS. Centrifuge cells for 5 min at 400 x g at 4 °C. After centrifugation, aspirate supernatant and resuspend pellet again in 5 ml of ice cold 3% FCS-PBS. Centrifuge cell suspensions for additional 5 min at 400 x g at 4 °C.
- Resuspend cells in an appropriate amount of EGM2 culture and proceed with step A4. Reduce light exposure during the subsequent steps and images (Figure 5) should be taken on an inverted microscope Eclipse TE2000-U (Nikon) or SteREO Lumar.V12 (Zeiss) over time. Here, a 20x objective was used for visualization. For each image, a phase contrast and fluorescence images in adequate channels (e.g., FITC-channel for CFSE labelled cells) were taken. SNARF-1 is normally used for detecting initial tube formation (a few hours after gelling). Later SNARF-1 labelling is difficult to detect. This system can be used to track the recruitment of perivascular cells to the endothelial cells during initial angiogenesis steps.
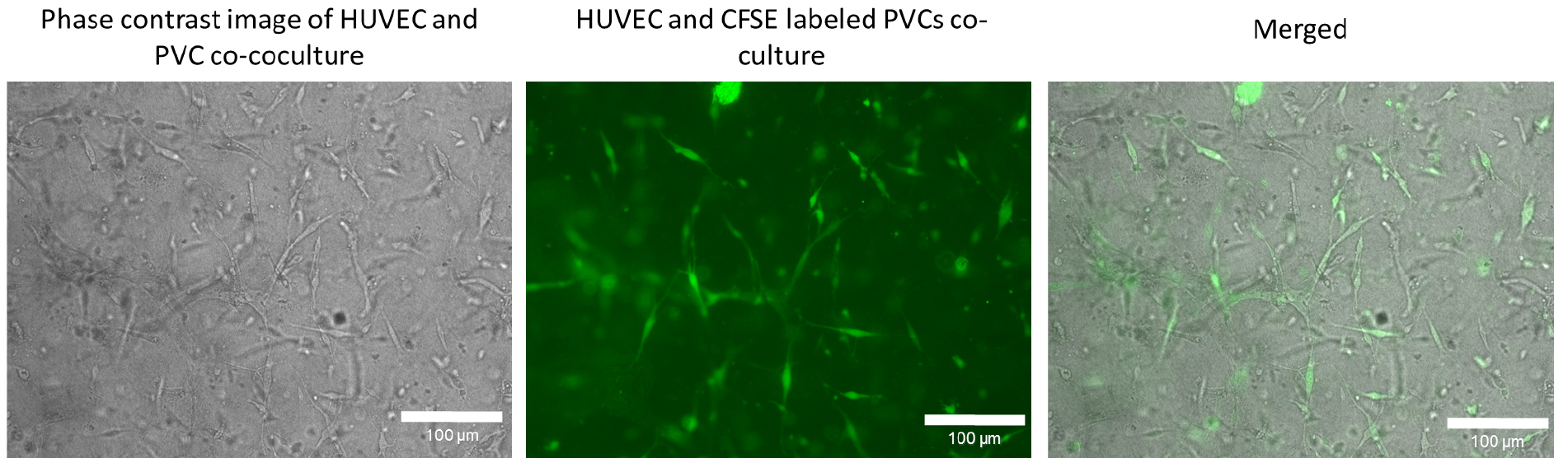
Figure 5. Co-culture of human umbilical cord vein endothelial cells (HUVEC) and CFSE labelled perivascular cells (PVC) to track perivascular cells in three-dimensional condition. PVCs were labelled with CFSE as described in Procedure D and subsequently co-cocultured with HUVECs in a three-dimensional collagen type I gel. After 72 h in culture, phase contrast image and fluorescence images were taken with a 20x objective. The brightness and contrast of the CFSE fluorescent and merged image were adjusted for visualization. Scale bars = 100 µm.
- Dissolve CFSE or SNARF-1 in DMSO to a final concentration of 5 mM at RT in the dark. After using fluorescent dyes, solution can be aliquoted and stored at -20 °C. For subsequent use of fluorescent dyes, thaw CFSE and SNARF-1 dye at RT in the dark. Freshly prepare 100 ml of 3% FCS-PBS solution. Store 25 ml at RT and cool the remaining 75 ml on ice for at least 30 min.
Data analysis
- The program ImageJ (Fiji) was used for image analysis.
- Open image file of interest (File → Open → Select image).
- Set scale. Choose ‘Arrow Tool’ in the toolbar and draw an arrow along the scale bar in the image. Next, click Analyze → Set Scale to open a new window, in which the known distance and unit of the length should be adjusted based on the distance and unit lengths acquired from the scale bar arrow. After, click ‘OK’.
- Go to toolbar and double click on ‘Arrow Tool’ to open a window. Choose a color for the arrow and click ‘keep after adding to overlay’. Next, click ‘OK’.
- Choose ‘Arrow Tool’ and draw an arrow along the tubes. Click Analyze → Measure (or STRG+M) to open a ‘Results’ window, which contains the length of this arrow. To add the arrow to the image, click Image → Overlay → Add Selection (or STRG+B).
- Draw an arrow along the next tube and measure the lengths as described in step D5.
- Once all the tubes have been measured, save the image with arrows (File → Save as → TIFF).
- Transfer the data from the ‘Results’ window into an Excel spreadsheet and determine the average of all measured tube lengths. It is recommended to acquire measurements from two or three, different images.
Notes
- Different endothelial cells (commercial providers) or perivascular cells isolated by various isolation procedures may be used for co-culture. In any case, cell numbers may have to be optimized for individual experiments.
- 100% confluent cultures should not be used for this assay, as it affects tube formation.
- Concentration of rat tail type I collagen can vary in different batches, volume have to be adjusted.
- Cell numbers that have not be adjusted accordingly can affect results.
- Include scale bar in each image for quantification.
- Carefully remove supernatant from gel without touching. Pipetting, rather than aspiration, is recommended for this step.
Recipes
- Proliferation medium
MCDB-131
5% FCS
10 ng/ml epidermal growth factor (EGF)
2 ng/ml basic fibroblast growth factor (bFGF)
5 µg/ml insulin
5 ng /ml platelet-derived growth factor-BB (PDGF-BB) - PVC culture medium (DMEM, 10% FCS, 1% P/S)
445 ml DMEM
50 ml FCS
5 ml P/S
Pre-warm medium before use at 37 °C - HUVEC culture medium (EGM2 with supplements)
500 ml basal medium
0.02 ml/ml FCS
5 ng/ml epidermal growth factor (EGF)
10 ng/ml basic fibroblast growth factor (bFGF)
20 ng/ml insulin-like growth factor (IGF)
0.5 ng/ml vascular endothelial growth factor (VEGF)
1 µg/ml ascorbic acid
22.5 µg/ml heparin
0.2 µg/ml hydrocortisone
Note: Basal medium should be ordered with supplement pack which includes all above listed supplements in the right concentration. Only small aliquots should be prewarmed at 37 °C. - 2x medium-mix (final volume 10 ml)
2 ml of 10x DMEM
0.2 ml of 100x sodium pyruvate
1 ml of 0.45 M NaHCO3
6.8 ml of autoclaved distilled H2O - Collagen solution (final volume 4 ml)
2 ml of 2x medium-mix
800 µl of FCS (final concentration 20%)
963 µl of 8.3 mg/ml rat tail type I collagen solution
214 µl of 0.1 N NaOH
23 µl of autoclaved distilled H2O
Prepare solution on ice and keep it on ice until usage - Tris-buffered saline (TBS), pH 7.4
150 mM NaCl
50 mM Tris-HCl
Acknowledgments
The three-dimensional co-culture assay was used in two recent publications (Pitzler et al., 2016; Zhou et al., 2016). The work is supported by Deutsche Forschungsgemeinschaft (DFG 2304/5-3, 2304/7-1, 2304/9-1).
References
- Baudin, B., Bruneel, A., Bosselut, N. and Vaubourdolle, M. (2007). A protocol for isolation and culture of human umbilical vein endothelial cells. Nat Protoc 2(3): 481-485.
- Brachvogel, B., Dikschas, J., Moch, H., Welzel, H., von der Mark, K., Hofmann, C. and Poschl, E. (2003). Annexin A5 is not essential for skeletal development. Mol Cell Biol 23(8): 2907-2913.
- Brachvogel, B., Moch, H., Pausch, F., Schlotzer-Schrehardt, U., Hofmann, C., Hallmann, R., von der Mark, K., Winkler, T. And Poschl, E. (2005). Perivascular cells expressing annexin A5 define a novel mesenchymal stem cell-like population with the capacity to differentiate into multiple mesenchymal lineages. Dev 132(11): 2657-2668.
- Brachvogel, B., Pausch, F., Farlie, P., Gaipl, U., Etich, J., Zhou, Z., Cameron, T., von der Mark, K., Bateman, J. F. and Poschl, E. (2007). Isolated Anxa5+/Sca-1+ perivascular cells from mouse meningeal vasculature retain their perivascular phenotype in vitro and in vivo. Exp Cell Res 313(12): 2730-2743.
- Pitzler, L., Auler, M., Probst, K., Frie, C., Bergmeier, V., Holzer, T., Belluoccio, D., van den Bergen, J., Etich, J., Ehlen, H., Zhou, Z., Bielke, W., Poschl, E., Paulsson, M. and Brachvogel, B. (2016). miR-126-3p promotes matrix-dependent perivascular cell attachment, migration and intercellular interaction. Stem Cells 34(5): 1297-1309.
- Zhou, Z., Pausch, F., Schlotzer-Schrehardt, U., Brachvogel, B. and Poschl, E. (2016). Erratum to: Induction of initial steps of angiogenic differentiation and maturation of endothelial cells by pericytes in vitro and the role of collagen IV. Histochem Cell Biol 145(5): 527-529.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Auler, M., Pitzler, L., Pöschl, E., Zhou, Z. and Brachvogel, B. (2017). Mimicking Angiogenesis in vitro: Three-dimensional Co-culture of Vascular Endothelial Cells and Perivascular Cells in Collagen Type I Gels. Bio-protocol 7(8): e2247. DOI: 10.21769/BioProtoc.2247.
Category
Stem Cell > Adult stem cell > Endothelial stem/progenitor cell
Cell Biology > Cell isolation and culture > Cell differentiation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









