- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Expression and Purification of Mini G Proteins from Escherichia coli
Published: Vol 7, Iss 8, Apr 20, 2017 DOI: 10.21769/BioProtoc.2235 Views: 13130
Reviewed by: Arsalan DaudiShyam SolankiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2171 Views

Prokaryotic Expression and Purification of the hSox2-HMG Domain
Lijie Yang [...] Jingjun Hong
Aug 20, 2025 2377 Views

On-Column Dual-Gradient Refolding for Efficient Recovery of Insoluble Affinity-Tagged Recombinant Proteins
Anna Vlaskina [...] Maxim Patrushev
Feb 5, 2026 66 Views
Abstract
Heterotrimeric G proteins modulate intracellular signalling by transducing information from cell surface G protein-coupled receptors (GPCRs) to cytoplasmic effector proteins. Structural and functional characterisation of GPCR–G protein complexes is important to fully decipher the mechanism of signal transduction. However, native G proteins are unstable and conformationally dynamic when coupled to a receptor. We therefore developed an engineered minimal G protein, mini-Gs, which formed a stable complex with GPCRs, and facilitated the crystallisation and structure determination of the human adenosine A2A receptor (A2AR) in its active conformation. Mini G proteins are potentially useful tools in a variety of applications, including characterising GPCR pharmacology, binding affinity and kinetic experiments, agonist drug discovery, and structure determination of GPCR–G protein complexes. Here, we describe a detailed protocol for the expression and purification of mini-Gs.
Keywords: ComplexBackground
We recently reported the development of an engineered minimal G protein, mini-Gs (Carpenter and Tate, 2016), which facilitated the crystallisation of the human adenosine A2A receptor (A2AR) in its active conformation (Carpenter et al., 2016; Carpenter and Tate, 2017). Unlike heterotrimeric G proteins, which require expression in eukaryotic systems, mini-Gs is highly expressed in Escherichia coli (E. coli) and can be easily purified with a yield of 50-100 mg of mini-Gs per liter of culture. Here, we describe a step by step protocol, earlier described in Carpenter and Tate (2016), that can be used for the expression and purification of any of the mini G protein constructs described previously (Carpenter et al., 2016; Carpenter and Tate, 2016). Since mini-Gs construct 393 is well suited to most applications (see Carpenter and Tate, 2016), it will be used as an example herein.
Materials and Reagents
- Steritop 0.22 μm filter unit (EMD Millipore, catalog number: SCGPT01RE )
- 50 ml tubes (SARSTEDT, catalog number: 62.547.254 )
- 15 ml tubes (SARSTEDT, catalog number: 62.554.002 )
- 2 ml tubes (Eppendorf, catalog number: 0030120094 )
- 1.5 ml tubes (SARSTEDT, catalog number: 72.690.001 )
- 0.5 ml tubes (SARSTEDT, catalog number: 72.699 )
- Pipette tips (STARLAB INTERNATIONAL)
- Plastic column (e.g., empty PD-10 column) (GE Healthcare, catalog number: 17043501 )
- HisTrap Fast Flow 5 ml prepacked columns (GE Healthcare, catalog number: 17-5255-01 )
- Amicon Ultra-15 concentrator 10 kDa cut-off (EMD Millipore, catalog number: UFC901024 )
- HiLoad 26/600 Superdex 200 PG gel filtration column (GE Healthcare, catalog number: 28989336 )
- SnakeSkin Dialysis Tubing 10 kDa cut-off (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 68100 )
- E. coli strain BL21-CodonPlus(DE3)-RIL (Agilent Technologies, catalog number: 230245 )
- pET15b plasmid (EMD Millipore, catalog number: 69661 )
- Ampicillin (Melford Laboratories, catalog number: A0104 )
- Chloramphenicol (MP Biomedicals, catalog number: 0219032125 )
- Glucose (Formedium, catalog number: GLU03 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (VWR, catalog number: 25165.260 )
- IPTG (Melford Laboratories, catalog number: MB1008 )
- Liquid nitrogen
- TEV protease (produced in-house)
- cOmplete, EDTA-free protease inhibitor tablets (Roche Diagnostics, catalog number: 11873580001 )
- Lysozyme (Sigma-Aldrich, catalog number: L6876 )
- Imidazole (Sigma-Aldrich, catalog number: 56748 )
- Ni2+-NTA agarose (QIAGEN, catalog number: 30210 )
- Gel filtration marker kit (Sigma-Aldrich, catalog number: MWGF200 )
- TCEP (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 77720 )
- Tryptone (Melford Laboratories, catalog number: GT1332 )
- Yeast extract (Melford Laboratories, catalog number: GY1333 )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: 10598630 )
- Agar
- Terrific Broth (TB) (Melford Laboratories, catalog number: GT1702 )
- Glycerol (VWR, catalog number: 24388.320 )
- PMSF (Sigma-Aldrich, catalog number: P7626 )
- Absolute ethanol (VWR, catalog number: 20821.330 )
- Pepstatin-A (Sigma-Aldrich, catalog number: P4265 )
- DMSO (Sigma-Aldrich, catalog number: D2650 )
- Leupeptin (Sigma-Aldrich, catalog number: L2884 )
- DNase I (Sigma-Aldrich, catalog number: DN25 )
- DTT (Melford Laboratories, catalog number: MB1015 )
- GDP (Sigma-Aldrich, catalog number: G7127 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- Magnesium chloride (MgCl2) (Fisher Scientific, catalog number: BP214-500 )
- Precision Plus SDS-PAGE molecular weight standards (Bio-Rad Laboratories, catalog number: 161-0373 )
- 4-20% Tris-Glycine SDS-PAGE gels (Fisher Scientific, catalog number: EC60255BOX )
- TYE agar plates (see Recipes)
- Luria Bertani (LB) media (see Recipes)
- Terrific Broth (TB) media (see Recipes)
- PMSF stock solution (see Recipes)
- Pepstatin-A stock solution (see Recipes)
- Leupeptin stock solution (see Recipes)
- DNase I stock solution (see Recipes)
- Lysozyme stock solution (see Recipes)
- DTT stock solution (see Recipes)
- GDP stock solution (see Recipes)
- Buffer A (see Recipes)
- Buffer B (see Recipes)
- Buffer C (see Recipes)
- Buffer D (see Recipes)
- Buffer E (see Recipes)
Equipment
- 2 L Erlenmeyer flasks (e.g., Corning, catalog number: 4980-2L )
- High speed centrifuge (e.g., Beckman Coulter, model: Avanti J-26XP , catalog number: 393124)
- Magnetic stirring bar
- Shaker incubator (Infors, model: Multitron Standard )
- Pipettes (STARLAB INTERNATIONAL)
- Sonicator equipped with 13 mm probe (e.g., Sonics Vibra-Cell) (Sonics, model: VCX 130 )
- Rotor capable of spinning 250 ml bottles (e.g., Beckman Coulter JLA-16.250) (Beckman Coulter, catalog number: 363934 )
- Peristaltic pump (e.g., GE Healthcare, model: Pump P-1, catalog number: 18-1110-91 )
- NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM 2000 )
- UV/VIS spectrophotometer
- Roller mixer (IKA, model: ROLLER 6 digital , catalog number: 0004011000)
- Rotor capable of spinning 1 L bottles (e.g., Beckman Coulter JLA-8.1000) (Beckman Coulter, catalog number: 363688 )
- Refrigerated benchtop centrifuge (e.g., Eppendorf, catalog number: 5430 R )
- Refrigerated microcentrifuge (e.g., Eppendorf, catalog number: 5418 R )
- ÄKTA Purifier chromatography system (GE Healthcare, model: ÄKTA Purifier )
Software
- UNICORN (GE Healthcare)
- Graphical software (e.g., Prism 7) (GraphPad), or free alternatives (e.g., R Bioconductor packages) (Bioconductor)
Procedure
- Expression of mini-Gs in E. coli
Mini-Gs393 was cloned into the pET15b plasmid to allow expression in E. coli. The construct contained an N-terminal histidine tag to facilitate purification of mini-Gs and a TEV protease cleavage site to allow removal of the histidine tag (Figure 1).
Figure 1. Mini-Gs393 E. coli expression construct. Mini-Gs was cloned into the plasmid pET15b (the sequence and map of this vector are available from the EMD Millipore website) using NcoI and XhoI restriction sites (underlined). The Mini-Gs393 construct contains an N-terminal 6x histidine tag (highlighted in red) followed by a TEV protease cleavage site (highlighted in green). Start and stop codons are shown in bold.
- Transform E. coli strain BL21-CodonPlus(DE3)-RIL with the plasmid pET15b-mini-Gs393, plate on a TYE agar plate (containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol) and incubate overnight at 37 °C. (see Note 1)
- Pick a single colony and inoculate 5 ml of LB media (supplemented with 0.2% glucose, 100 μg/ml ampicillin and 34 μg/ml chloramphenicol), incubate for 6-8 h at 37 °C, shaking at 220 rpm.
- Inoculate 150 ml of LB media (supplemented with 0.2% glucose, 100 μg/ml ampicillin and 34 μg/ml chloramphenicol) with 5 ml of the starter culture, incubate for 16-20 h at 30 °C, shaking at 220 rpm.
- Measure the OD600 nm of the overnight culture and inoculate 4 x 500 ml of TB media (supplemented with 0.2% glucose, 100 μg/ml ampicillin, 34 μg/ml chloramphenicol and 5 mM MgSO4) in 2 L Erlenmeyer flasks to give an OD600 nm of 0.15
- Incubate at 30 °C, shaking at 220 rpm, until the OD600 nm reaches 0.6-0.8 (this should take 2-3 h). Induce expression of mini-Gs by addition of IPTG to give a final concentration of 50 μM, incubate at 25 °C, shaking at 220 rpm for 16-20 h.
- Harvest the cells by centrifugation at 5,000 x g for 10 min at 4 °C. Flash freeze the pellet in liquid nitrogen and store at -80 °C.
- Transform E. coli strain BL21-CodonPlus(DE3)-RIL with the plasmid pET15b-mini-Gs393, plate on a TYE agar plate (containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol) and incubate overnight at 37 °C. (see Note 1)
- Purification of mini-Gs
Mini-Gs393 is purified by Ni2+ affinity chromatography, followed by cleavage of the histidine tag using TEV protease and negative purification on Ni2+-NTA to remove the TEV and undigested mini-Gs. A final gel filtration step is performed to remove aggregated protein. This protocol yields approximately 100 mg of pure mini-Gs393 from 2 L of E. coli culture.- Resuspend the pellet from 2 L of E. coli culture in buffer A to give a final volume of 200 ml.
- Add 4 protease inhibitor tablets, and PMSF (1 mM), Pepstatin-A (2.5 μM), Leupeptin (10 μM), DNase I (50 μg/ml), lysozyme (50 μg/ml) and DTT (100 μM) to give the final concentrations indicated.
- Stir the cell suspension at medium speed using a magnetic stirring bar at 4 °C for 30 min.
- Lyse the cells by sonication for 10 min, with pulses of 2 sec on and 4 sec off, at an amplitude of 70%, keeping the sample in an ice bath to prevent heating.
- Transfer the lysate to a 250 ml centrifuge bottle and centrifuge at 38,000 x g for 45 min at 4 °C.
- Remove the supernatant and filter through a 0.22 μm Steritop filter unit. (see Note 2)
- Load the supernatant onto a 10 ml HisTrap Fast Flow column (2 x 5 ml columns connected in series), pre-equilibrated with buffer A, at a flow rate of 5 ml/min at 4 °C, using a peristaltic pump.
- Wash the column with 100 ml of buffer B at a flow rate of 5 ml/min at 4 °C.
- Elute the column with 30 ml of buffer C at a flow rate of 5 ml/min at 4 °C, collecting a single 30 ml fraction.
- Determine the concentration of mini-Gs by measuring the A280 nm using a NanoDrop 2000 spectrophotometer. An absorbance of 1.0 unit equates to a protein concentration of 1.0 mg/ml for mini-Gs393 (extinction coefficient: 27,310 M-1 cm-1).
- A maximum of 200 mg of protein can be processed from this point onwards, if more mini-Gs is present it can be split into two batches and processed in parallel. (see Note 3)
- Add DTT to give a final concentration of 1 mM and TEV protease to give a TEV:mini-Gs ratio of 1:20 w/w. (see Note 4)
- Dialyse the protein against 1 L of buffer D for 2-3 h at 4 °C, discard the external buffer and dialyse against 1 L of fresh buffer D overnight at 4 °C.
- Transfer the protein to a 50 ml tube, add imidazole to give a final concentration of 20 mM and 4 ml of Ni2+-NTA agarose resin (pre-equilibrated with buffer D), mix on a roller mixer for 30 min at 4 °C. (see Note 5)
- Pour the suspension onto 1 ml of Ni2+-NTA agarose resin (pre-equilibrated with buffer D) packed into a disposable plastic column and let it run through by gravity.
- Collect the flow through and wash the column with 5 ml of buffer D.
- Pool the wash with the flow through and concentrate to 1.5 ml using an Amicon Ultra-15 concentrator (10 kDa cut-off) in a refrigerated benchtop centrifuge at 4 °C (this may take up to 2 h).
- Centrifuge the concentrated protein at > 15,000 x g for 10 min in a refrigerated microcentrifuge at 4 °C to remove aggregates.
- Load the supernatant onto a HiLoad 26/600 Superdex 200 gel filtration column (pre-equilibrated with buffer E) at a flow rate of 2.6 ml/min at 4 °C, collecting 2 ml fractions. (see Note 6)
- Pool peak fractions (typically 22-26 ml) based on the gel filtration chromatogram (Figure 2).
- Concentrate the pooled fractions to 100 mg/ml, using an Amicon Ultra-15 concentrator (10 kDa cut-off) in a refrigerated benchtop centrifuge at 4 °C (this may take up to 2 h).
- Determine the concentration of mini-Gs by measuring the A280 nm using a NanoDrop 2000 spectrophotometer. To ensure an accurate reading, dilute a sample of the protein 1:10 in buffer E before measuring the absorbance.
- Centrifuge the concentrated protein at > 15,000 x g for 10 min in a refrigerated microcentrifuge at 4 °C to remove aggregates.
- Transfer the supernatant to a fresh tube and discard the pellet.
- Aliquot 20 μl of the supernatant (2 mg of mini-Gs) into 0.5 ml tubes, flash freeze in liquid nitrogen, store at -80 °C. Mini-Gs is stable under these conditions for over 1 year.
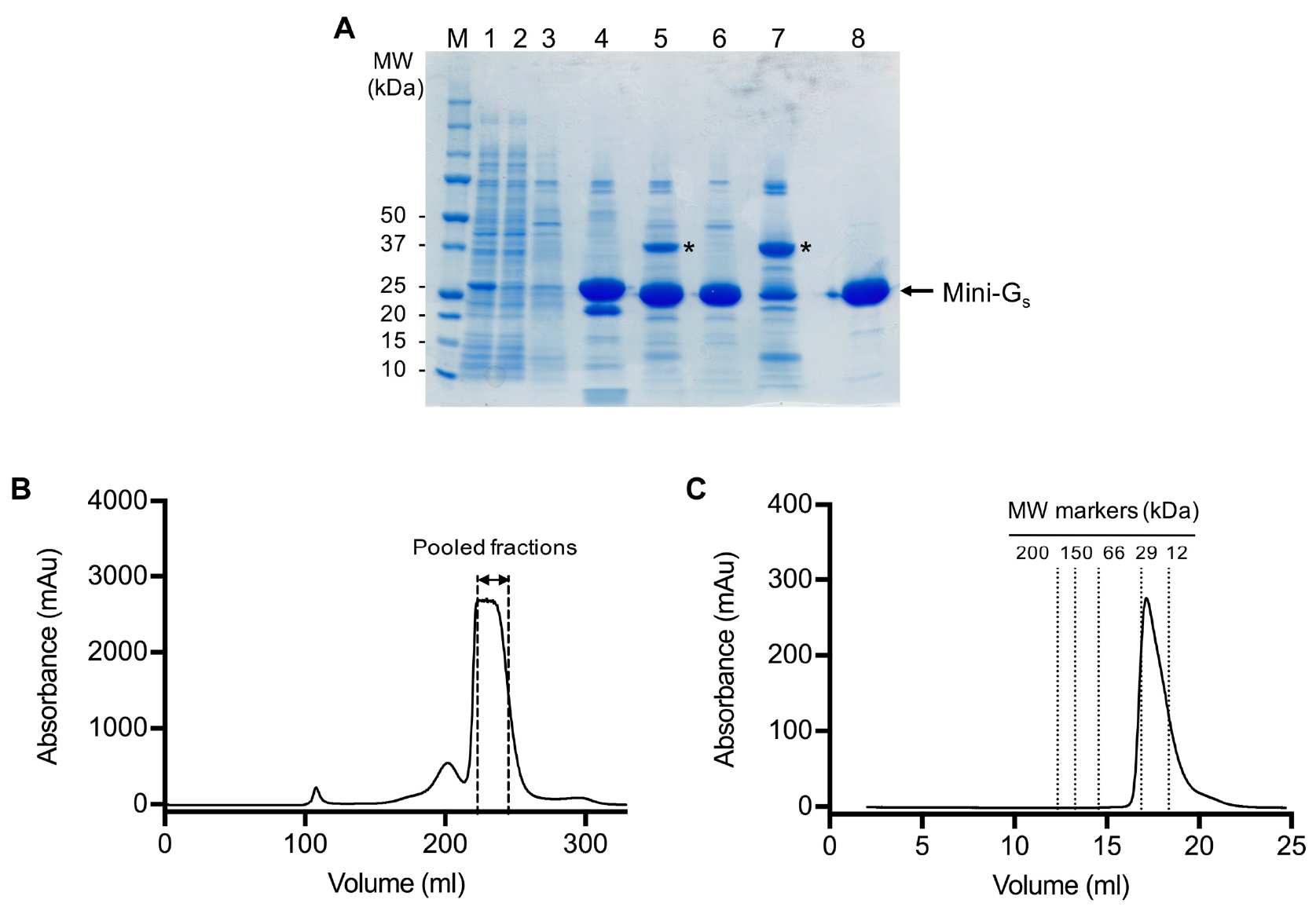
Figure 2. SDS-PAGE and gel filtration analysis of the mini-Gs393 purification. A. SDS-PAGE analysis of the mini-Gs393 purification. Lane: (M) molecular weight markers; (1) HisTrap column loading material (1:10 dilution); (2) HisTrap column flow through (1:10 dilution); (3) HisTrap column wash; (4) HisTrap column eluate; (5) sample after TEV digestion; (6) Ni2+-NTA negative purification flow through; (7) Ni2+-NTA negative purification eluate; (8) gel filtration pool. TEV protease is indicated by an asterisk. Note that it is not necessary to boil the samples before loading on the gel. B. Preparative gel filtration chromatogram for mini-Gs393. Pooled fractions are indicated by dashed lines, fractions at the leading edge of the peak should be omitted to ensure that aggregates are completely removed. Note that the UV detector saturates at absorbance readings above 2,700 mAu, which gives the chromatogram a truncated absorbance profile. C. Analytical gel filtration analysis of purified mini-Gs on a Superdex 200 GL 10/300 column. The chromatogram demonstrates that purified mini-Gs is monomeric (calculated molecular weight of 23 kDa compared to the theoretical value of 27 kDa) and free from aggregates.
- Resuspend the pellet from 2 L of E. coli culture in buffer A to give a final volume of 200 ml.
Data analysis
Chromatograms were recorded using UNICORN software and data were plotted using GraphPad Prism 7. The analytical gel filtration column was calibrated using molecular weight standards (gel filtration marker kit), and the apparent molecular weight of mini-Gs was calculated from the calibration curve as previously published (Carpenter and Tate, 2016). No statistical analysis was performed during this work.
Notes
- The BL21-CodonPlus(DE3)-RIL E. coli strain contains a plasmid, carrying the chloramphenicol resistance gene, which encodes extra copies of rare tRNAs. This helps to negate the effect of codon bias when expressing eukaryotic proteins in E. coli. The standard BL21(DE3) strain can also be used for expression of mini-Gs, but in this case chloramphenicol should be omitted from all media.
- Filtration of the lysate significantly extends the life of the HisTrap column, which can be regenerated and reused at least 10 times.
- The maximum loading capacity of the HiLoad 26/600 Superdex 200 gel filtration column is approximately 150 mg of protein. TEV digestion of 200 mg of protein from the HisTrap eluate will yield approximately 150 mg of mini-Gs after the Ni2+-NTA negative purification step.
- TEV protease is available from commercial suppliers such as Sigma-Aldrich, but can be expensive. Alternatively, it can be easily purified in-house using any one of the protocols reported in the literature (for example, Tropea et al., 2009). The main consideration is that the purified TEV should contain a polyhistidine tag to facilitate its removal by Ni2+ negative purification after cleavage of mini-Gs.
- Mini-Gs has significant affinity for Ni2+ resin even after cleavage of the histidine tag. It is vital to add 20 mM imidazole at this step, otherwise mini-Gs will bind to the Ni2+ resin and will not be recovered in the flow through. This is also an important consideration for any downstream application where purified mini-Gs may contact Ni2+ resin, for example pull-down experiments.
- Mini-Gs tends to aggregate through nonspecific disulphide bonds, therefore TCEP is added to buffer E to maintain a reducing environment. DTT can be used instead of TCEP, but is less stable in solution.
Recipes
- TYE agar plates
10 g/L tryptone
5 g/L yeast extract
8 g/L NaCl
15 g/L agar
Autoclave at 121 °C, 2.8 bar for 15 min to sterilise. Add ampicillin (100 μg/ml) and chloramphenicol (34 μg/ml) to give the final concentrations indicated - Luria-Bertani (LB) media
10 g/L tryptone
5 g/L yeast extract
10 g/L NaCl
Autoclave at 121 °C, 2.8 bar for 15 min to sterilise - Terrific Broth (TB) media
24 g/L yeast extract
12 g/L tryptone
9.4 g/L K2HPO4
2.2 g/L KH2PO4
4 ml/L glycerol
Autoclave at 121 °C, 2.8 bar for 15 min to sterilise - PMSF stock solution
200 mM in absolute ethanol, store at 4 °C - Pepstatin-A stock solution
2.5 mM in DMSO, store at -20 °C - Leupeptin stock solution
10 mM in Milli-Q water, store at -20 °C - DNase I stock solution
50 mg/ml in Milli-Q water, store at -20 °C - Lysozyme Lysozyme stock solution
50 mg/ml in Milli-Q water, store at -20 °C - DTT stock solution
1 M in Milli-Q water, make fresh - GDP stock solution
100 mM in Milli-Q water, make fresh - Buffer A
40 mM HEPES, pH 7.5
100 mM NaCl
10 mM imidazole
10% v/v glycerol
5 mM MgCl2
50 μM GDP (add immediately before use)
Filter using a 0.22 μm Steritop or syringe filter - Buffer B
20 mM HEPES, pH 7.5
500 mM NaCl
40 mM imidazole
10% v/v glycerol
1 mM MgCl2
50 μM GDP (add immediately before use)
Filter using a 0.22 μm Steritop or syringe filter - Buffer C
20 mM HEPES, pH 7.5
100 mM NaCl
500 mM imidazole
10% v/v glycerol
1 mM MgCl2
50 μM GDP (add immediately before use)
Filter using a 0.22 μm Steritop or syringe filter - Buffer D
20 mM HEPES, pH 7.5
100 mM NaCl
10% v/v glycerol
1 mM MgCl2
10 μM GDP (add immediately before use) - Buffer E
10 mM HEPES, pH 7.5
100 mM NaCl
10% v/v glycerol
1 mM MgCl2
1 μM GDP (add immediately before use)
0.1mM TCEP (add immediately before use)
Filter using a 0.22 μm Steritop or syringe filter
Acknowledgments
This work was funded by the Wellcome Trust, Heptares Therapeutics Ltd and core funding from the Medical Research Council [MRC U105197215]. This protocol was adapted from a previously described method (Carpenter and Tate, 2016). We thank Rony Nehmé for comments on the manuscript.
References
- Carpenter, B., Nehmé, R., Warne, T., Leslie, A. G. and Tate, C. G. (2016). Structure of the adenosine A2A receptor bound to an engineered G protein. Nature 536(7614): 104-7.
- Carpenter, B. and Tate, C. G. (2016). Engineering a minimal G protein to facilitate crystallisation of G protein-coupled receptors in their active conformation. Protein Eng Des Sel 29(12): 583-594.
- Carpenter, B. and Tate, C.G. (2017). Expression, purification and crystallisation of the adenosine A2A receptor bound to an engineered mini G protein. Bio-protocol 7(8): e2234.
- Tropea, J.E., Cherry, S. and Waugh, D.S. (2009). Expression and purification of soluble His6-tagged TEV protease. Methods Mol Biol 498: 297-307.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Carpenter, B. and Tate, C. G. (2017). Expression and Purification of Mini G Proteins from Escherichia coli. Bio-protocol 7(8): e2235. DOI: 10.21769/BioProtoc.2235.
Category
Biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Expression
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











