- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Rapid Isolation of Total Protein from Arabidopsis Pollen
Published: Vol 7, Iss 8, Apr 20, 2017 DOI: 10.21769/BioProtoc.2227 Views: 11228
Reviewed by: Marisa RosaYuan ChenMohan TCIgor Cesarino

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2869 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1989 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1772 Views
Abstract
Arabidopsis pollen is an excellent system for answering important biological questions about the establishment and maintenance of cellular polarity and polar cell growth, because these processes are amenable to the genetic and genomic approaches that are readily available in Arabidopsis. Given that proteins are the direct executors of a wide variety of cellular processes, it is important to rapidly and efficiently isolate total protein for various protein-based analyses, such as Western blotting, co-immunoprecipitation and mass spectrometry, among others. Here we present a protocol for rapid isolation of total protein from Arabidopsis pollen, which is adapted from our recently published paper (Chang and Huang, 2015).
Keywords: Arabidopsis thalianaBackground
Pollen is a critical stage during the life cycle of sexually-reproducing plants. Pollen germination and subsequent tube growth provide the passage for two non-motile sperm cells to effect double fertilization in flowering plants. Pollen is routinely used as a model system to addressing fundamental cell biological questions, such as the establishment and maintenance of cell polarity and polar cell growth, as well as the structure and function of the actin cytoskeleton (Chen et al., 2009; Qu et al., 2015). Generally, pollen derived from lily and tobacco was widely used due to the large size of the grains, which makes them easy to collect and observe under the microscope. In contrast, Arabidopsis pollen is small and it is relatively difficult to collect a large amount of pollen to isolate a sufficient quantity of protein for downstream analyses such as Western blotting and mass spectrometry. Therefore, development of a protocol to rapidly isolate total protein from Arabidopsis pollen will facilitate related analyses.
Materials and Reagents
- 1.5 ml Eppendorf tubes (Fisher Scientific, FisherbrandTM, catalog number: 05-408-129 )
- Pipette tips (USA Scientific, catalog numbers: 1112-1720 , 1110-1200 )
- Arabidopsis plants (ecotype Col-0)
- Ponceau S (Sigma-Aldrich, catalog number: P3504 )
- Anti-α-tubulin (Beyotime Biotechnology, catalog number: AT819 )
- Anti-actin (Abmart, catalog number: M20009 )
- Anti-COXII (Agrisera, catalog number: AS04 053A )
- 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (AMRESCO, catalog number: 0511 )
- K-acetate (Sinopharm Chemical Reagents, catalog number: 30154518 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma, catalog number: 63064 )
- Tween 20 (AMRESCO, catalog number: 0777 )
- Triton X-100 (Sigma-Aldrich, catalog number: V900502 )
- Phenylmethanesulfonyl fluoride (PMSF) (EMD Millipore, catalog number: 52332 )
- K-HEPES protein extraction buffer (see Recipes)
Equipment
- Pestles (VWR, catalog number: 89093-446 )
- Forceps (Shanghai Haiou)
- Pipettes (VWR, catalog numbers: 89079-970 , 89079-974 )
- Vortex (Corning, model: Corning® LSETM Vortex Mixer )
- Centrifuge (Eppendorf, model: 5417 R )
Procedure
- Grow Arabidopsis plants in a growth room at 21 °C under long-day conditions (16 h light/8 h dark) for 6 to 8 weeks. Normally, the Col-0 ecotype will produce many flowers under these conditions. The plant growth time can be changed to adapt to the flowering time of other Arabidopsis ecotypes.
- Quickly collect more than 100 freshly opened flowers (stage 13-15, Figure 1) in one 1.5 ml Eppendorf tube and add 1 ml K-HEPES protein extraction buffer (see Recipes). This step can be done at room temperature.
Note: Collect Arabidopsis flowers around 10 AM. At this time the flowers will be fully opened, allowing easier access to the pollen.
Figure 1. An opening Arabidopsis flower at stage 13-15 (Müller, 1961; Smyth et al., 1990) - Vortex the flowers at the highest speed for 2-5 min in order to thoroughly separate the pollen from the flowers (Video 1).Video 1. Procedure for the isolation of pollen from flowers by vortex in step 3
- Centrifuge at 350 x g for 1 min at 4 °C (Video 2).
Note: Under low speed, most of the flowers will be suspended in the upper layer of buffer, but the pollen will be centrifuged to the bottom of the tube.Video 2. Procedure for the isolation of protein as stated in step 4-7 - Slowly and thoroughly remove all the flower debris with small forceps (Video 2).
Note: If necessary, you can repeat steps 4 and 5 several times for total removal of the flower debris, but you should be careful to avoid touching the pollen at the bottom of the tube. - Completely remove the supernatant (Video 2).
Note: Make sure the pollen is still at the bottom of the tube. - Put the tube on ice and thoroughly grind the pollen with a small pestle for 1-3 min (Video 2). The liquid mixture will turn yellow color if the pollen is homogenized completely.
Note: If your lab doesn’t have suitable pestles for 1.5 ml tubes, you can use a 1 ml pipette tip, sealed by melting it in a flame, as a substitute. - Add 50 μl K-HEPES protein extraction buffer into the tube and keep the tube on ice for about 1 h.
- Centrifuge at 18,000 x g for 20 min at 4 °C, then transfer the supernatant to a new 1.5 ml tube (keep the tube on ice). Repeat this step twice. The supernatant is the final pollen protein sample and the protein concentration can be measured by standard Bradford assay.
Data analysis
SDS-PAGE and Western blot analyses of Arabidopsis pollen total proteins are shown in Figure 2. Total proteins isolated from Arabidopsis rosette leaves, in which the small subunit of Rubisco is by far the most abundant component, were used as the control (Figure 2A). In contrast, the band corresponding to the small subunit of Rubisco is hardly detected in the pollen total proteins (Figure 2B). This means that detection of proteins other than the small subunit of Rubisco might be easier in Arabidopsis total protein extract. For instance, by loading 10 μg of total proteins from pollen per lane, α-tubulin (TUA), actin (ACT), cytochrome c oxidase subunit II (COXII) and profilin (PRF) can be easily detected (Figure 2B).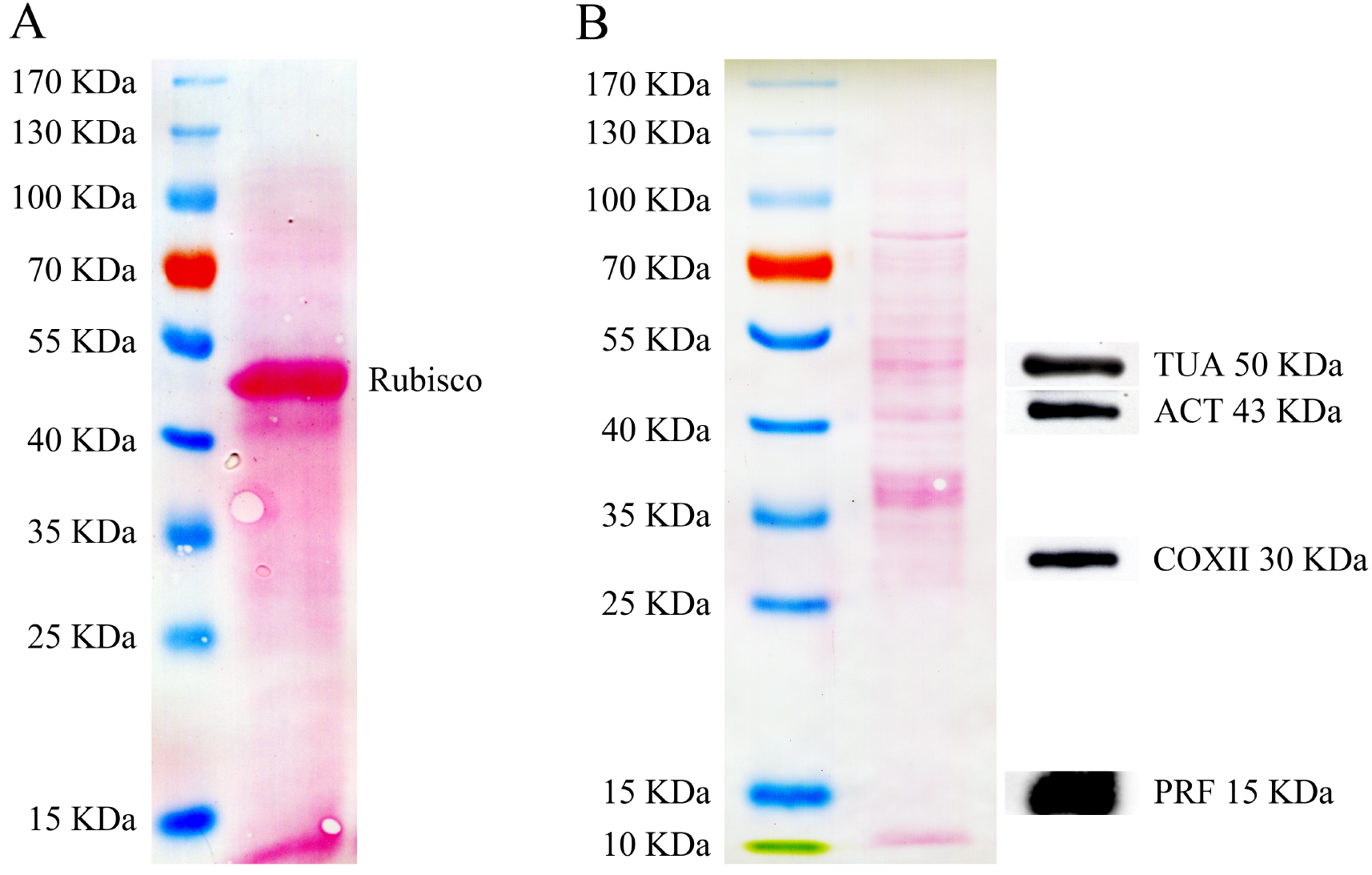
Figure 2. SDS-PAGE analysis of total proteins isolated from Arabidopsis pollen and leaves, and Western blot analysis of total proteins from Arabidopsis pollen probed with several antibodies. A. SDS-PAGE analysis of total proteins (50 μg) isolated from Arabidopsis rosette leaves. The nitrocellulose (NC) membrane was stained with Ponceau S after protein transfer. B. The left panel shows the SDS-PAGE analysis of total proteins (10 μg) isolated from Arabidopsis pollen. The NC membrane was stained with Ponceau S after protein transfer. The right panel shows the Western blot analysis of Arabidopsis pollen total proteins probed with anti-α-tubulin, anti-actin, anti-COXII and profilin antibodies. The profilin antibody was generated in our laboratory; see the details in our published paper (Liu et al., 2015). Other antibodies are commercially available: anti-α-tubulin (Beyotime Biotechnology, AT819), anti-actin (Abmart, M20009) and anti-COXII (Agrisera, AS04 053A).
Notes
Generally, 100 Col-0 flowers will yield about 100 μg total protein from pollen with this method.
Recipes
- K-HEPES protein extraction buffer
20 mM HEPES, pH 7.0
110 mM K-acetate
2 mM MgCl2
0.1% Tween 20
0.2% Triton X-100
1 mM PMSF (add immediately before using)
Note: This buffer (without the PMSF) can be stored for up to 6 months at 4 °C.
Acknowledgments
This protocol was modified from our previously published work (Chang and Huang, 2015). The work in Huang’s lab was supported by funding from the Ministry of Science and Technology of China (2013CB945100) and the National Natural Science Foundation of China (31671390 and 31471266).
References
- Chang, M. and Huang, S. (2015). Arabidopsis ACT11 modifies actin turnover to promote pollen germination and maintain the normal rate of tube growth. Plant J 83(3): 515-527.
- Chen, N., Qu, X., Wu, Y. and Huang, S. (2009). Regulation of actin dynamics in pollen tubes: control of actin polymer level. J Integr Plant Biol 51(8): 740-750.
- Liu, X., Qu, X., Jiang, Y., Chang, M., Zhang, R., Wu, Y., Fu, Y. and Huang, S. (2015). Profilin regulates apical actin polymerization to control polarized pollen tube growth. Mol Plant 8(12): 1694-1709.
- Müller, A. (1961). Zur Charakterisierung der Blüten und Infloreszenzen von Arabidopsis thaliana (L.) Heynh. Kulturpflanze 9(1): 364-393.
- Qu, X., Jiang, Y., Chang, M., Liu, X., Zhang, R. and Huang, S. (2015). Organization and regulation of the actin cytoskeleton in the pollen tube. Front Plant Sci 5: 786.
- Smyth, D. R., Bowman, J. L. and Meyerowitz, E. M. (1990). Early flower development in Arabidopsis. Plant Cell 2(8): 755-767.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Chang, M. and Huang, S. (2017). Rapid Isolation of Total Protein from Arabidopsis Pollen. Bio-protocol 7(8): e2227. DOI: 10.21769/BioProtoc.2227.
Category
Plant Science > Plant biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











