- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Microtubule Bundling Assay under Physiological Conditions
Published: Vol 7, Iss 7, Apr 5, 2017 DOI: 10.21769/BioProtoc.2217 Views: 9514
Reviewed by: Jyotiska ChaudhuriRebecca Van AckerElizabeth Libby

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Rapid and solvent-free, 2-hydroxyethyl methacrylate (HEMA)-acrylamide (AAm) copolymer-based optical clearing of tissue for fluorescent imaging
Yanran Wang [...] Kefeng Wu
Nov 20, 2025 1516 Views

Characterizing Tissue Oxygen Tension During Neurogenesis in Human Cerebral Organoids
Yuan-Hsuan Liu and Hsiao-Mei Wu
Nov 20, 2025 1715 Views

SiMPull-POP: Quantification of Membrane Protein Assembly via Single Molecule Photobleaching
Ryan J. Schuck [...] Rajan Lamichhane
Jan 5, 2026 304 Views
Abstract
Kinesins play a role in organizing the mitotic spindle through the crosslinking of microtubules (MTs), made possible through binding sites at opposite ends of the holoenzyme. Here, we developed a method to test kinesin MT crosslinking action under physiological conditions.
Keywords: in vitro bundling assayBackground
Microtubule-based motor proteins are important as they use the chemical energy from ATP to generate force to translocate microtubules (MTs) vectorially. Of these MT based motor proteins, the superfamily known as kinesins, are responsible for directional transport and movement along microtubules. Some kinesins have MT-binding sites at both ends of the holoenzyme, so they can crosslink MTs into bundles under physiological ATP conditions (Tao et al., 2006). Due to this bundling activity, they have important roles in organizing and maintaining the mitotic spindle, whose action depends upon the polarity patterns of its microtubules (van den Wildenberg et al., 2008). Previous bundling assays didn’t include ATP, or used a non-hydrolysable ATP analog, which could generate artificial results. Here we developed a method using physiological ATP conditions. By purifying these full-length motor proteins, it has allowed us to determine their crosslinking activity under physiological conditions.
Materials and Reagents
- Microcentrifuge tubes (1.5 ml) (Sigma-Aldrich, catalog number: Z336769 )
- Aluminum foil
- Double sided tape
- Coverslip, No. 1.5-22 x 22 mm (Sigma-Aldrich, catalog number: Z692263 )
- Microscope slides (AmScope, catalog number: BS-50P )
- Kimwipe (Sigma-Aldrich, catalog number: Z188956 )
- Pipette tips (Corning, catalog number: 4860 )
- Tubulin (CYTOSKELETON, catalog number: MT001-A )
- Glycerol (Sigma-Aldrich, catalog number: G2025 )
- Rhodamine tubulin (CYTOSKELETON, catalog number: TL590M-A )
- GTP (Sigma-Aldrich, catalog number: G8877 )
- Hydrochloric acid (HCl [37%, v/v]) (Sigma-Aldrich, catalog number: 320331 )
- Full-length kinesin motor proteins are expressed and purified from baculoviral expression system
- ATP (Sigma-Aldrich, catalog number: A6419 )
- Piperazine-N,N’-bis(2-ethanesulfonic acid) (PIPES) (Sigma-Aldrich, catalog number: P6757 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M4880 )
- EGTA (Sigma-Aldrich, catalog number: 03777 )
- Potassium hydroxide (KOH)
- Paclitaxel (Sigma-Aldrich, catalog number: T7402 )
- DMSO (Sigma-Aldrich, catalog number: D2650 )
- DEAE-dextran (Sigma-Aldrich, catalog number: 30461 )
- Tris base (Sigma-Aldrich, catalog number: T1503 )
- Potassium chloride (KCl) (Sigma-Aldrich: P5405 )
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D0632 )
- Protease inhibitors
Aprotinin (Sigma-Aldrich, catalog number: A3428 )
Benzamidine (Sigma-Aldrich, catalog number: B6505 or 12072 )
Note: The product “ B6505 ” has been discontinued.
Pepstatin A (Sigma-Aldrich, catalog number: P5318 )
Phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626 )
Leupeptin (Sigma-Aldrich, catalog number: L2884 )
Soy Bean Trypsin inhibitor (SBTI) (Sigma-Aldrich, catalog number: T6522 )
tert-Amyl methyl ether (TAME) (Sigma-Aldrich, catalog number: 283096 ) - Catalase (Sigma-Aldrich, catalog number: C1345 )
- Glucose oxidase (Sigma-Aldrich, catalog number: C6766 or G7141 )
Note: The product “ C6766 ” has been discontinued. - Glucose (Sigma-Aldrich, catalog number: G7021 or D9434 )
- 10x phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: P5493 )
- BRB80 (see Recipes)
- 1 µM, 10 µM, and 100 µM taxol (see Recipes)
- DEAE-dextran (see Recipes)
- Buffer T (see Recipes)
- Antifade (see Recipes)
Equipment
- Nichipet EX-Plus II pipette 2-20 μl (Nichiryo, catalog number: 00-NPLO2-20 )
- Nichipet EX-Plus II pipette 10-100 μl (Nichiryo, catalog number: 00-NPLO2-100 )
- Water bath, from ambient to 100 °C (Thermo Fisher Scientific, Thermo ScientificTM, model: PrecisionTM General Purpose Baths , catalog number: TSGP02)
- Flow chamber (see Procedure)
- Inverted fluorescence microscope (12-volt 100-watt tungsten-halide lamp, excitation wavelength, 547 nm; emission wavelength, 576 nm, in conjunction with a 570 nm dichroic mirror, objective: CFI Plan Fluor 100x oil) (Nikon Instruments, model: Eclipse E600 )
Procedure
- Polymerization of fluorescent MTs
- In a 1.5 ml microcentrifuge tube, dissolve 0.25 mg tubulin in 60 µl of BRB80 solution (containing 10% glycerol). Store on ice.
- Add 20 µg of Rhodamine tubulin into the 60 µl solution. Mix gently for 30 sec and store tube half submerged in an ice bucket.
- Add 1/10 the total volume of the 60 µl of tubulin mix, worth of 10 mM GTP, to the 60 µl tubulin mix, with a final concentration of 1 mM GTP. Mix gently by inverting.
- Add 1/10 the total volume of mixture step 1c, worth of 1 µM taxol, to the mix, with a final taxol concentration of 0.1 µM. Incubate (in water bath) at 37 °C for 10 min.
- Add 1/10 the total volume of mixture step 1d, worth of 10 µM taxol, to the mix, with a final taxol concentration of 1 µM. Incubate (in water bath) at 37 °C for 10 min.
- Add 1/10 the total volume of mixture step 1e, worth of 100 µM taxol, to the mix, with a final taxol concentration of 10 µM. Incubate (in water bath) at 37 °C for 10 min.
- Store the polymerized MTs at room temperature, with the tube covered in aluminum foil and stored in a closet to protect the mixture from light. The MTs are stable for use for up to two weeks.
- Prepare chamber
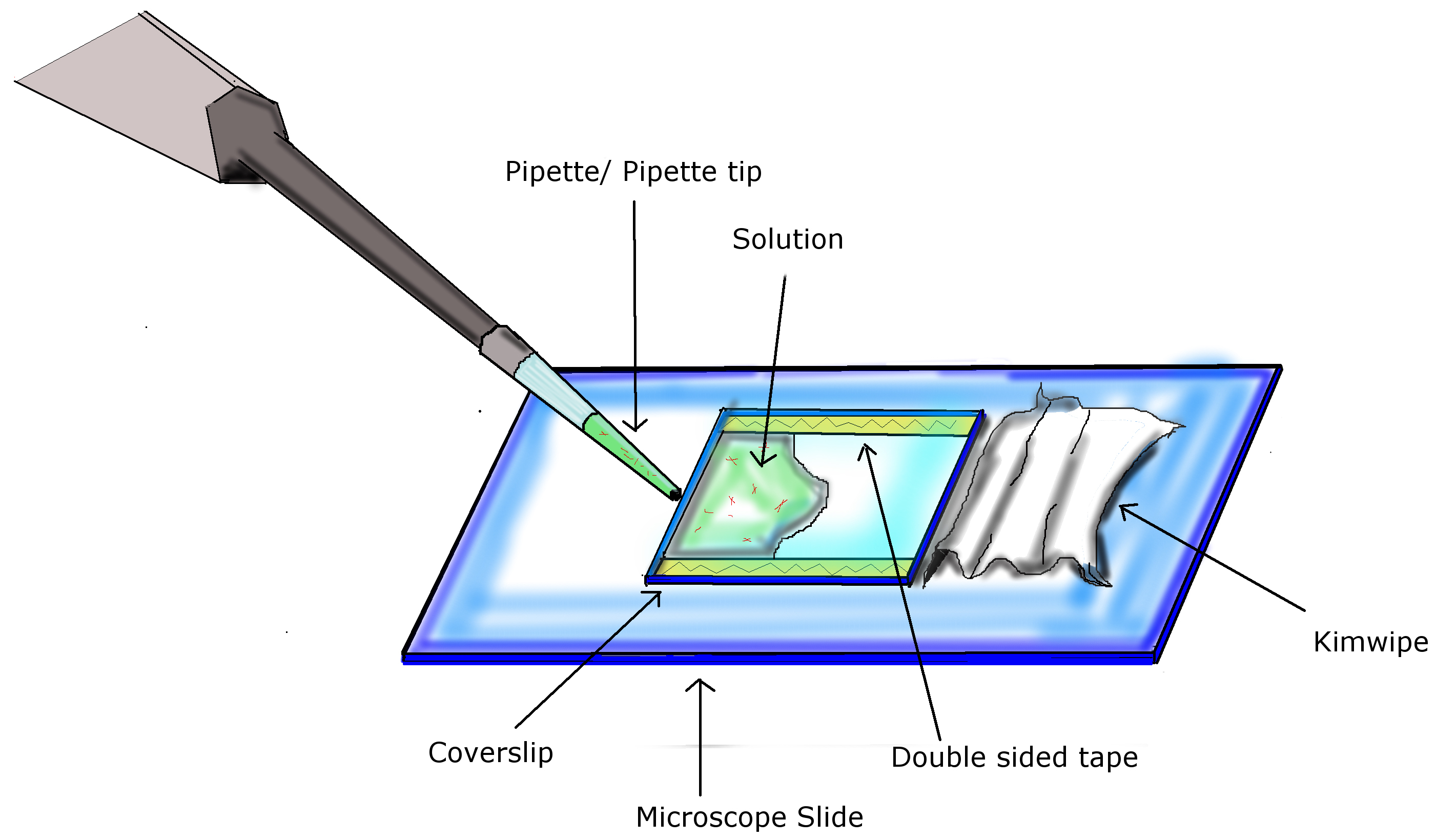
Figure 1. Infusion of chamber. Using two pieces of double sided tape, a coverslip is attached to a microscope slide, completing the preparation of the flow chamber. Using a pipette with appropriate volumes, infuse chamber by pipetting solution into the opening of the flow chamber. A Kimwipe is used to facilitate diffusion of the solution across the chamber and absorb excess liquid at the end. - Acid wash coverslip with HCl (37%, v/v) for 24 h, rinse with de-ionized water and air dry for 24 h.
- After cleaning, put a drop of 0.2 mg/ml DEAE-dextran on coverslip and let sit for 5 min.
- Absorb remaining liquid with Kimwipe and air dry coverslip.
- Place two strips of double sided tape (L x 22 mm) parallel to each other on microscope slide at a distance of 22 mm from each other. Place the coverslip down, matching the alignment of the tape, with the DEAE-dextran coated side facing towards the microscope slide. Refer to Figure 1.
- Prepare bundling mixture.
- Prepare 25 µl mix in buffer T, with the following reagents to a final concentration of; 2.5 mM MTs, 0.2 mM motor proteins, 10 µM taxol, and 1 mM ATP.
Note: For motor proteins, we use hydrodynamic assays (Tao et al., 2016) to determine the MW. For MT, we use MW of tubulin dimer (α tubulin + β tubulin) to calculate the concentration of MTs. - Shake the mixture at 50 rpm at room temperature for 30 min.
- Infuse mix from step 3 into flow chamber:
- Infuse 25 µl into the flow chamber(s), let sit for 3 min. Refer to Figure 1.
- Wash out any unstuck proteins with a continuous flow of 25 µl buffer T solution (including reagents with a final concentration of 1 mM ATP, 10 mM taxol, and Antifade), repeated two additional times. Use a similar technique as infusion. Refer to Figure 1.
- Observe and record MT bundling using inverted fluorescence microscope (12-volt 100-watt tungsten-halide lamp, excitation wavelength, 547 nm; emission wavelength, 576 nm in conjunction with a 570 nm dichroic mirror, objective: CFI Plan Fluor 100x oil, Nikon Instruments, model: Eclipse E600).
Data analysis
The microtubule (MT) bundling assay was repeated three times, with three separate batches of reagents. 15 images were collected for each experiment. To ensure consistency, it is recommended that the assay be repeated three times, with 15 images collected for each. Figure 2 shows a kinesin holoenzyme crosslinking MTs into bundles. Figure 2A shows a robust bundle due to crosslinking activity of the kinesin motor. However, in Figure 2B the kinesin-1 head dimers do not bundle MTs, suggesting that the binding sites at opposite ends of the motor are required for crosslinking, rather than action of two heads at one end.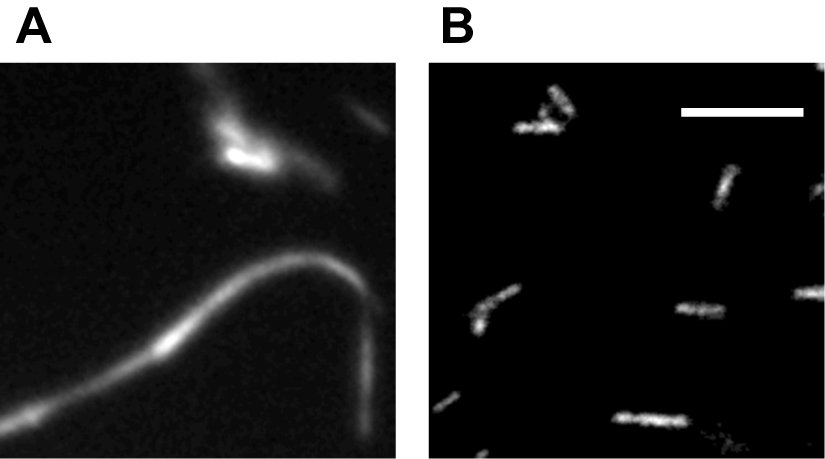
Figure 2. Example of bundling assay. Fluorescence microscopy of purified kinesin holoenzyme bundling MTs (A) while kinesin-1 head dimers unable to bundle MTs under same condition (B). Scale bar = 10 µm. (Tao et al., 2016)
Notes
- To ensure maximum bundling activity, purify the motor proteins within 30 min before use in bundling assay.
- For best results, fluorescent microtubules should be used within two weeks of their polymerization, with the mixture being stored in aluminum foil covered container, kept in a dark location.
- Full-length kinesin motor cDNA was cloned into a Gateway Baculovirous expression vector (pDEST8) with a 6xHis tag. After verification by sequencing, the recombinant expression vector was used to generate recombinant Baculoviruses. Baculovirus-infected Sf9 cells were collected and then recombinant motor protein was purified on a Ni-NTA column using standard protocol (Tao et al., 2010).
Recipes
- BRB80*
80 mM PIPES
1 mM MgCl2
1 mM EGTA
Adjust pH to 6.8 with KOH - 1 µM, 10 µM, and 100 µM taxol
20 µl of 1 mM taxol dissolved in 180 µl DMSO = 200 µl of 100 µM taxol
6 µl of 100 µM taxol dissolved in 54 µl DMSO = 60 µl of 10 µM taxol
6 µl of 10µM taxol dissolved in 54 µl DMSO = 60 µl of 1 µM taxol - DEAE-dextran
0.2 mg/ml of ddH2O - Buffer T*
20 mM Tris
150 mM KCl
2 mM MgCl2
1 mM DTT
Adjust pH to 8.0 with KOH
Protease inhibitors with final concentration of Aprotinin 2 µg/ml, Benzamidine 20 µg/ml, Pepstatin A 1 µg/ml, PMSF 0.1 mM, Leupeptin 1 µg/ml, SBTI 100 µg/ml, Tame 40 µg/ml, added just before use - Antifade
10 mg catalase
3 mg glucose oxidase
180 mg glucose
100 µl 1 M DTT
1.90 mg EGTA
100 µl 10x PBS
Fill to 1 ml with ddH2O
*Note: Listed concentrations are final concentrations, buffer is prepared in ddH2O.
Acknowledgments
This MT bundling assay was inspired by ‘Tum/RacGAP functions as a switch activating the Pav/kinesin-6 motor’ (Tao et al., 2016) and ‘Purification and assay of mitotic motors’ (Tao and Scholey, 2010). This work was funded by the NIH grant GM046409 to W.S. and NIH grant GM 55507 to J.M.S.
References
- Tao, L., Fasulo, B., Warecki, B. and Sullivan, W. (2016). Tum/RacGAP functions as a switch activating the Pav/kinesin-6 motor. Nat Commun 7: 11182.
- Tao, L., Mogilner, A., Civelekoglu-Scholey, G., Wollman, R., Evans, J., Stahlberg, H. and Scholey, J. M. (2006). A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol 16(23): 2293-2302.
- Tao, L. and Scholey, J. M. (2010). Purification and assay of mitotic motors. Methods 51(2): 233-241.
- van den Wildenberg, S. M., Tao, L., Kapitein, L. C., Schmidt, C. F., Scholey, J. M. and Peterman, E. J. (2008). The homotetrameric kinesin-5 KLP61F preferentially crosslinks microtubules into antiparallel orientations. Curr Biol 18(23): 1860-1864.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Geisterfer, Z., Tezuka, G. and Tao, L. (2017). In vitro Microtubule Bundling Assay under Physiological Conditions. Bio-protocol 7(7): e2217. DOI: 10.21769/BioProtoc.2217.
Category
Cell Biology > Cell imaging > Fluorescence
Biochemistry > Protein > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









