- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Gliding Assay to Analyze Microtubule-based Motor Protein Dynamics
Published: Vol 7, Iss 7, Apr 5, 2017 DOI: 10.21769/BioProtoc.2210 Views: 14398
Reviewed by: Jyotiska ChaudhuriTamara VellosilloJason A. Neidleman

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Rapid and solvent-free, 2-hydroxyethyl methacrylate (HEMA)-acrylamide (AAm) copolymer-based optical clearing of tissue for fluorescent imaging
Yanran Wang [...] Kefeng Wu
Nov 20, 2025 1516 Views

Characterizing Tissue Oxygen Tension During Neurogenesis in Human Cerebral Organoids
Yuan-Hsuan Liu and Hsiao-Mei Wu
Nov 20, 2025 1715 Views

SiMPull-POP: Quantification of Membrane Protein Assembly via Single Molecule Photobleaching
Ryan J. Schuck [...] Rajan Lamichhane
Jan 5, 2026 303 Views
Abstract
The purpose of this protocol is to provide an updated method of performing microtubule gliding assays and visualizing it using fluorescence microscopy.
Keywords: Gliding assayBackground
Mitotic spindles are protein machinery that dominate mitosis. The mitotic spindle utilizes microtubule-based motor proteins to organize itself, and exert forces to drive cell division. Microtubule-based motor proteins produce mechanical work using energy derived from ATP hydrolysis (Coppin et al., 1997). Motor proteins translocate microtubules in a unidirectional manner. The behavior of motility can be observed by in vitro gliding assay (Tao and Scholey, 2010), in which the motors are affixed onto a glass surface and supplied with microtubules and ATP. The motility of the motor proteins can then be studied using fluorescence microscopy and the details of their dynamic behavior can be observed in real time. This updated protocol will allow analysis of microtubule-based motor protein function with the use of in vitro microtubule gliding assays (Tao et al., 2006 and 2016).
Materials and Reagents
- Pipette tips (Corning, catalog number: 4860 )
- Centrifuge tubes (Corning, catalog number: 430290 )
- Thickwall polycarbonate tubes (Beckman Coulter, catalog number: 343775 )
- Coverslips (Sigma-Aldrich, catalog number: Z692263 )
- Microscope slides (Fisher Scientific, catalog number: S17466A )
- Coverslips (Fisher Scientific, catalog number: S175211A )
- Guanosine-5’-[(α,β)-methyleno]triphosphate (GMPCPP) (10 mM) (Jena Bioscience, catalog number: NU-405S )
- Tubulin (CYTOSKELETON, catalog number: TL238A )
- Rhodamine tubulin (CYTOSKELETON, catalog number: TL590M )
- Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D9779 )
- N-ethylmaleimide (Sigma-Aldrich, catalog number: E3876 )
- Guanosine 5’-triphosphate (GTP) (Sigma-Aldrich, catalog number: G8877 )
- Hydrogen chloride (HCl) (Sigma-Aldrich, catalog number: 295426 )
- Casein (Sigma-Aldrich, catalog number: C7078 )
- ATP (Sigma-Aldrich, catalog number: A6419 )
- Paclitaxel (Taxol) (Sigma-Aldrich, catalog number: T7402 )
- Piperazine-N,N’-bis(2-ethanesulfonic acid) (PIPES) (Sigma-Aldrich, catalog number: P6757 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M4880 )
- EGTA (Sigma-Aldrich, catalog number: E3889 )
- Tris base (Sigma-Aldrich, catalog number: T1503 )
- Potassium chloride (KCl) (Sigma-Aldrich: P5405 )
- Protease inhibitors:
Aprotinin (Sigma-Aldrich, catalog number: A3428 )
Benzamidine (Sigma-Aldrich, catalog number: B6505 or 12072 )
Note: The product “ B6505 ” has been discontinued.
Pepstatin A (Sigma-Aldrich, catalog number: P5318 )
Phenylmethylsulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626 )
Leupeptin (Sigma-Aldrich, catalog number: L2884 )
Soybean trypsin inhibitor (SBTI) (Sigma-Aldrich, catalog number: T6522 )
Tert-Amyl methyl ether (TAME) (Sigma-Aldrich, catalog number: 283096 ) - Catalase (Sigma-Aldrich, catalog number: C1345 )
- Glucose oxidase (Sigma-Aldrich, catalog number: C6766 or G7141 )
Note: The product “ C6766 ” has been discontinued. - Glucose (Sigma-Aldrich, catalog number: D9434 )
- Phosphate buffered saline (PBS) (Sigma-Aldrich, catalog number: P5368 )
- BRB80 buffer solution (see Recipes)
- Buffer L (see Recipes)
- Anti-fade solution (see Recipes)
Equipment
- Micropipette
2-20 μl (Sigma-Aldrich, catalog number: Z717304 )
10-100 μl (Sigma-Aldrich, catalog number: Z717312 ) - Ultra-centrifuge (Beckman Coulter, model: TLA-100 )
- NanoDrop (Thermo Fisher Scientific, Thermo ScientificTM, model: NanoDropTM 2000 )
- Fluorescence microscope (Nikon Instruments, model: Eclipse E600 )
- Water bath (Thermo Fisher Scientific, model: PrecisionTM General Purpose Baths , catalog number: TSGP02)
Software
- ImageJ (Version 1.51J, https://imagej.nih.gov/ij/)
Procedure
- Polarity marked microtubules
- Make bright GMPCPP seed, which serves as a ‘primer’ for microtubule polymerization as well as an indicator of the microtubule minus-end.
- Make a 10 mg/ml solution of tubulin by dissolving 250 µg of tubulin into 25 µl of BRB80.
- Make a 10 mg/ml solution of rhodamine tubulin by dissolving 20 µg into 2 µl of BRB80.
- Mix 4 µl of the 10 mg/ml tubulin solution, 2 µl of the10 mg/ml rhodamine tubulin solution, 18 µl of BRB80, 3 µl of 10 mM DTT, and 3 µl 10 mM GMPCPP. Store mix on ice.
- Centrifuge at 410,000 x g for 7 min at 4 °C.
- Take out 3 µl of bright seed mix supernatant and incubate in water bath at 37 °C for 15 min.
- Make N-ethylmaleimide tubulin
- Make a 10 mg/ml tubulin solution on ice (see step A1a).
- Make a fresh 1 mM N-ethylmaleimide (NEM) solution.
Add 0.5 μl 50 mM NEM to 25 μl 10 mg/ml tubulin. Let the solution sit at room temperature for 5 min. - Add 0.5 µl of 1 M DTT to 25 µl NEM-tubulin solution. Let the solution sit on ice for 30 min.
- Centrifuge NEM-tubulin solution at 350,000 x g for 15 min at 4 °C. Collect the supernatant.
- Measure the concentration of supernatant with NanoDrop at 280 nm absorbance.
- Make dim elongation mix
- To make 10 μl elongation mix:
1.34 μl unlabeled tubulin (10 mg/ml)
0.17 μl rhodamine tubulin (10 mg/ml)
3 μl NEM-tubulin (4 mg/ml)
1 μl 10 mM DTT (diluted 1 M DTT into 10 mM DTT first)
0.2 μl 50 mM GTP
4.29 μl 1x BRB80 - Incubate the mix in water bath at 37 °C for 1 min.
- Make polarity marked microtubules.
- Add 2 µl of bright seed mix to 10 µl dim elongation mix and gently mix.
- Incubate the solution in water bath at 37 °C for 30 min, and add taxol stepwise to 10 µM final solution. Store the microtubules at room temperature and protect from light.
- Gliding assay
- Make glass chamber
- Acid wash coverslips (22 x 22 mm) with HCl for 24 h and rinse with ddH2O for 10 min each, 3 times. Air dry the coverslips for 24 h.
- Attach the coverslip to a glass slide (75 x 25 mm) using two pieces of double-sided tape on the top and bottom. There should be open slits on both sides (Figure 1).
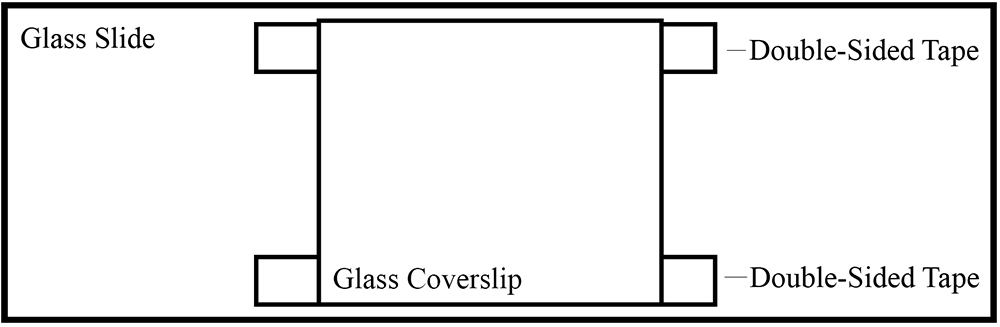
Figure 1. Illustration of glass slide chamber using double-sided tape - Flow in 25 µl of 1 mg/ml casein into the chamber and let it sit for 5 min. Casein serves as a cushion to anchor motor proteins.
- Wash out the unbinding casein with 3 x 20 µl buffer L.
- Flow in 10-100 nM purified motor protein (in buffer L), and let it sit for 5 min. Full-length kinesin motor proteins are expressed and purified from baculoviral expression system.
- Wash out unbinding proteins with 3 x 20 µl buffer L.
- Flow in 25 µl polarity marked microtubules at a 1/250 dilution into the chamber.
For 100 µl volume, mix 95.4 µl buffer L, 1 µl 100 mM ATP, 1 µl 1 mM taxol, 2 µl anti-fade, 0.2 µl 1 M DTT, and 0.4 µl MT’s.
- Acid wash coverslips (22 x 22 mm) with HCl for 24 h and rinse with ddH2O for 10 min each, 3 times. Air dry the coverslips for 24 h.
- Observe microtubule motility under fluorescence microscope (Nikon E600, excitation wavelength, 547 nm; emission wavelength, 576 nm. Also, see following Video 1).Video 1. Example of microtubule gliding assay
- Make glass chamber
Data analysis
- Figure 2 illustrates the principle of a gliding assay. The motor proteins are fixed onto the coverslip and then an aliquot of fluorescent microtubules is introduced into the chamber and caught by the motor. Individual fluorescent microtubules move continuously in a unidirectional manner. If the motor is plus-end directed, the microtubule itself will translocate in the minus-end leading direction, and vice versa for a minus-end directed motor.
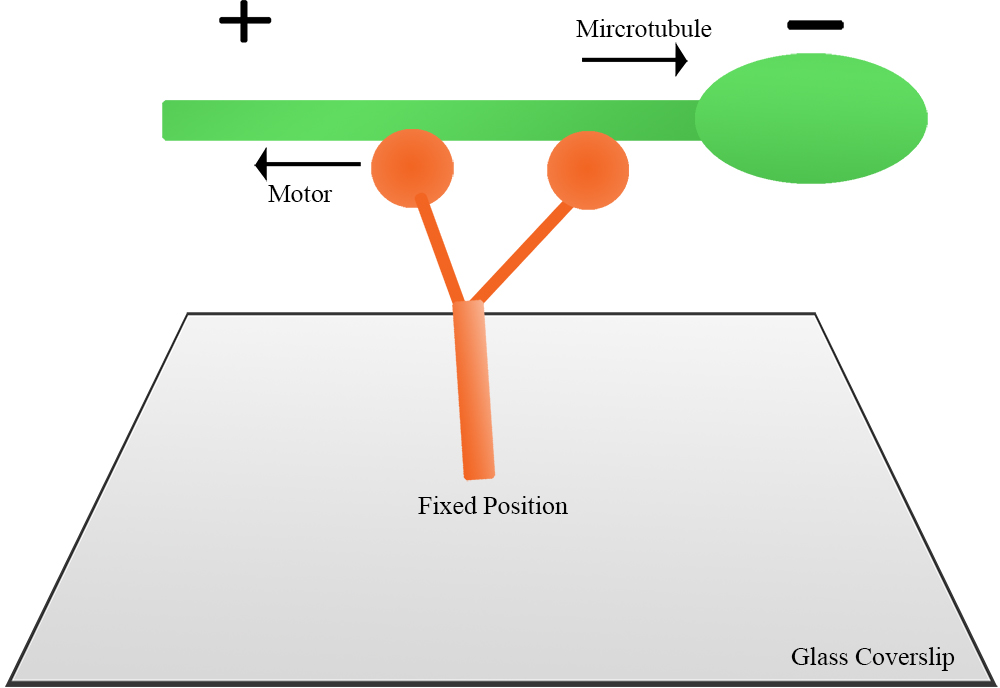
Figure 2. Illustration of a gliding assay. Full-length motor protein is fixed onto coverslip, then polarity-marked microtubules are introduced to the chamber. Microtubule motility is observed under fluorescence microscope. - Figure 3 shows a plus-end directed motor moving microtubules. The microtubule is in the minus-end leading direction. ImageJ was used to measure the distance of the microtubule movement, and the velocity was subsequently calculated.

Figure 3. Polarity-marked microtubule translocation in a gliding assay. MT gliding driven by purified kinesin-5 protein. MT is minus-end leading, which suggests that kinesin-5 is a plus-end-directed motor. Scale bar = 5 µm (Tao et al., 2006).
Notes
- In the gliding assay, the motor proteins must be freshly purified to ensure maximum activity.
- While making polarity marked microtubules, it is recommended to prepare the bright GMPCPP seed and the dim elongation mix simultaneously. Bright seed mix needs to be added to dim elongation mix right after the seed is made.
Recipes
- BRB80 buffer solution
1.80 mM PIPES
1 mM MgCl2
1 mM EGTA, pH 6.8 - Buffer L
1.20 mM Tris
75 mM KCl
2 Mm MgCl2
1 mM DTT
0.1 mM ATP
Adjust pH to 8.0
Protease inhibitors (Final concentrations: Aprotinin, 2 μg/ml; Benzamidine, 20 μg/ml; Leupeptin, 1 μg/ml; Pepstatin A, 1 μg/ml; PMSF, 0.1 mM; SBTI, 100 μg/ml; TAME, 40 μg/ml), added just before use - Anti-fade solution
- Mix 10 mg of catalase, 3 mg of glucose oxidase, 180.2 mg glucose, 100 µl 1 M DTT, 1.902 mg EGTA, and 100 µl 10x PBS
- Fill with ddH2O up to 1 ml
- Aliquot into 20 µl, and store at -80 °C
Acknowledgments
This method is adopted from Tao et al. (2016) and Tao et al. (2010). The above work was funded by NIH grant GM046409 to W.S. and NIH grant GM 55507 to J.M.S.
References
- Coppin, C. M., Pierce, D. W., Hsu, L. and Vale, R. D. (1997). The load dependence of kinesin's mechanical cycle. Proc Natl Acad Sci U S A 94(16): 8539-8544.
- Tao, L., Fasulo, B., Warecki, B. and Sullivan, W. (2016). Tum/RacGAP functions as a switch activating the Pav/kinesin-6 motor. Nat Commun 7: 11182.
- Tao, L., Mogilner, A., Civelekoglu-Scholey, G., Wollman, R., Evans, J., Stahlberg, H. and Scholey, J. M. (2006). A homotetrameric kinesin-5, KLP61F, bundles microtubules and antagonizes Ncd in motility assays. Curr Biol 16(23): 2293-2302.
- Tao, L. and Scholey, J. M. (2010). Purification and assay of mitotic motors. Methods 51(2): 233-241.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Shim, A., Tezuka, G., Kupcha, L. and Tao, L. (2017). Gliding Assay to Analyze Microtubule-based Motor Protein Dynamics. Bio-protocol 7(7): e2210. DOI: 10.21769/BioProtoc.2210.
Category
Cell Biology > Cell imaging > Fluorescence
Biochemistry > Protein > Fluorescence
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










