- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of the Galactanase Activity of the GanB Galactanase Protein from Bacillus subtilis
Published: Vol 7, Iss 7, Apr 5, 2017 DOI: 10.21769/BioProtoc.2206 Views: 8959
Reviewed by: Valentine V TrotterPetru-Iulian TrasneaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Surface Plasmon Resonance for the Interaction of Capsular Polysaccharide (CPS) With KpACE
Zhe Wang [...] Chao Cai
Jun 20, 2025 3554 Views

Thermus thermophilus CRISPR Cas6 Heterologous Expression and Purification
Junwei Wei [...] Yingjun Li
Jul 20, 2025 2171 Views

An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics
Saurabh Upadhyay
Aug 20, 2025 2455 Views
Abstract
The activity of the endo-β-1,4-galactanase GanB from B. subtilis on the high molecular weight β-1,4-galactan was determined quantitatively by the measurement of the increase of the reducing power or with the dyed substrate Azo-galactan. The generated degradation products were analyzed using thinlayer-chromatography (TLC) or high-performance anion-exchange chromatography (HPAEC).
Keywords: Galactanase-assayBackground
Bacillus subtilis possesses comprehensive systems for the utilization of plant cell wall polysaccharides including the gene cluster ganSPQAB which encodes galactan utilization elements (Watzlawick et al., 2016). Galactans are high molecular weight galacto-saccharides and are found as side chains of rhamnogalacturonan type I in pectin. Its degradation is carried out by endo-beta1,4 galactanases (EC 3.2.1.89). The utilization of galactan by B. subtilis involves the extracellular galactanase GanB, cleaving the high molecular galactan inside the chain and resulting short oligomers that enter the cell wall to get there further degraded. The ganB gene from B. subtilis was cloned and expressed in E.coli (Watzlawick et al., 2016) and the enzymatic properties of the purified His-tagged mature protein were characterized by galactanase assays.
Materials and Reagents
- Eppendorf tubes 1.5 ml
- TLC-plate: Silica Gel 60 F254 (Merck Millipore, Darmstadt, Germany)
- Sodium acetate trihydrate (Carl Roth, catalog number: 6779 )
- Galactan from lupin (Arabinofuranosidase treated lupin pectic galactan, Gal:Ara:Rha:Xyl: GalUA = 91:2:1.8:0.2:5.0) (Megazyme, catalog number: P-GALLU )
- β-1,4-galactobiose (Sigma-Aldrich, catalog number: G9662 )
- D-(+)-galactose (Sigma-Aldrich, catalog number: G5388 )
- 3,5-dinitrosalicylic acid (Sigma-Aldrich, catalog number: D0550 )
- Potassium-sodium-tartrate tetrahydrate (Carl Roth, catalog number: 8087 )
- Sodium sulfite (Carl Roth, catalog number: P033 )
- Sodium hydroxide 50% (Carl Roth, catalog number: 8655 )
- AZCL-galactan: Azurine-crosslinked-potato (AZCL) galactan (which has been purified from potato fibre and treated to remove most of the arabino- furanosyl residues) (Sigma-Aldrich, catalog number: 38127 )
Note: This product has been discontinued. - Ammonium heptamolybdate tetrahydrate (Carl Roth, catalog number: 7311 )
- Cerium sulfate tetrahydrate (Carl Roth, catalog number: P014 )
- Sulfuric acid (H2SO4)
- Potassium-phosphate dibasic (KH2PO4) (Carl Roth, catalog number: P018 )
- di-Potassium hydrogen phosphate (K2HPO4) (Carl Roth, catalog number: P749 )
Note: Prepare a 0.1 M solution in pure H2O and use it for preparation of the potassium phosphate buffer described in Recipes section. - Sodium borate decahydrate (Sigma-Aldrich, catalog number: G5388 )
- Sulfuric acid 95% (Carl Roth, catalog number: HN62 )
- Acetone (Carl Roth, catalog number: 9372 )
- Milli-Q pure H2O
- Purified recombinant His6-tagged GanB (see Recipes)
Note: This protein was produced in E. coli JM109 harboring the plasmid pHWG1119 after induction with rhamnose as described by (Watzlawick et al., 2016). Crude extracts were purified by immobilized-metal affinity chromatography using TALON® Metal Affinity resins (Clontech Laboratories, USA) according to the supplier’s instruction. Fractions of the purified proteins were combined and the imidazole was removed with NAP10 columns (GE Healthcare, Munich, Germany) equilibrated with 0.1 M KPP. - Lupin-galactan (see Recipes)
- DNS-reagent (see Recipes)
- AZCL-galactan (see Recipes)
- TLC developing reagent (see Recipes)
- Potassium phosphate buffer (KPP), 0.1 M (see Recipes)
- 0.4 M sodium borate solution (see Recipes)
- 0.3 M sodium acetate, pH 5.2 (see Recipes)
- TLC mobile phase solution (see Recipes)
- HPAEC isocratic mobile phase (see Recipes)
Equipment
- Pipette P2 , P20 , P200 , P1000 (Gilson Pipetman classic)
- Vortex Minishaker (Heidolph Instrument, model: Heidolph Reax 2000 )
- Centrifuge (Eppendorf, model: 5415 D )
- Thermomixer compact (Eppendorf)
- Blow-dryer
- HPLC apparatus: The HPLC apparatus consists of a pump (220, Bischoff, Leonberg, Germany) and an ESA Coulochem II electrochemical detector (Bischoff, Leonberg, Germany) with an analytical cell (5040, ESA Coulochem II, Bischoff)
- HPLC-column (Thermo Fisher Scientific, model: DionexTM CarboPacTM PA1 )
- TLC-chamber (size: 12 x 12 x 7 cm) (Desaga, Heidelberg, Germany)
- Spectrophotometer (Thermo Fisher Scientific, Thermo ScientificTM, model: GENESYSTM 10 Vis )
- pH meter (Metteler Toledo, model: FE20 ATC FiveEasyTM )
Procedure
In order to measure the galactanase activity of His6-GanB two different assays were applied: one using the natural substrate galactan is described in Procedure A and another using a dyed galactan substrate is described in Procedure B. The reaction of the galactanase enzyme on galactan for producing galacto-oligosaccharide is described in Procedure C. Galacto-oligosaccharides generated after the degradation of galactan were identified by two different chromatographic methods. First Thinlayer-chromatography (TLC) was used to identify the products qualitatively and is described in Procedure D. Quantitative analysis of individual galacto-oligosaccharides was performed using high-performance anion-exchange chromatography (HPAEC) as described in Procedure E.
- Galactanase activity determination by measurement of the increase of the reducing power
In order to calculate endo-β-galactanase activity in units, the reduced sugars released from the lupin galactan degradation were determined by an adapted 3,5-dinitrosalicylic acid (DNS) assay (Miller, 1959).- Mix appropriate amounts of His6-GanB (maximal 0.5 µg) in 40 µl of 0.1 M potassium phosphate buffer (KPP) pH 6.5 with 40 µl of 2% lupin galactan in an Eppendorf tube.
- Incubate the Eppendorf tube in a thermomixer at 37 °C for 15 min with agitation at 300 rpm.
- Stop the reaction by adding of 300 µl of DNS-reagent.
- Heat the Eppendorf tube for 20 min at 95 °C.
- Add 1 ml H2O to the reaction mixture and keep the Eppendorf tube on ice for 20 min.
- Measure the absorbance of the developed orange color at 540 nm (see Figure1)
As the blank the reaction mixture ingredients without enzyme is used.
For unit calculation, the absorption of 1 µmol galactose is determined from a calibration curve applying 1 to 10 mM galactose in the reaction mixture of the assay (1 µmol galactose is equivalent to 0.034 A540 units). One unit of galactanase enzyme was defined as the release of 1 µmol of reducing sugar, here galactose, per minute determined by the DNS-assay. Calculate units per milliliter from the formula (see also Data analysis):
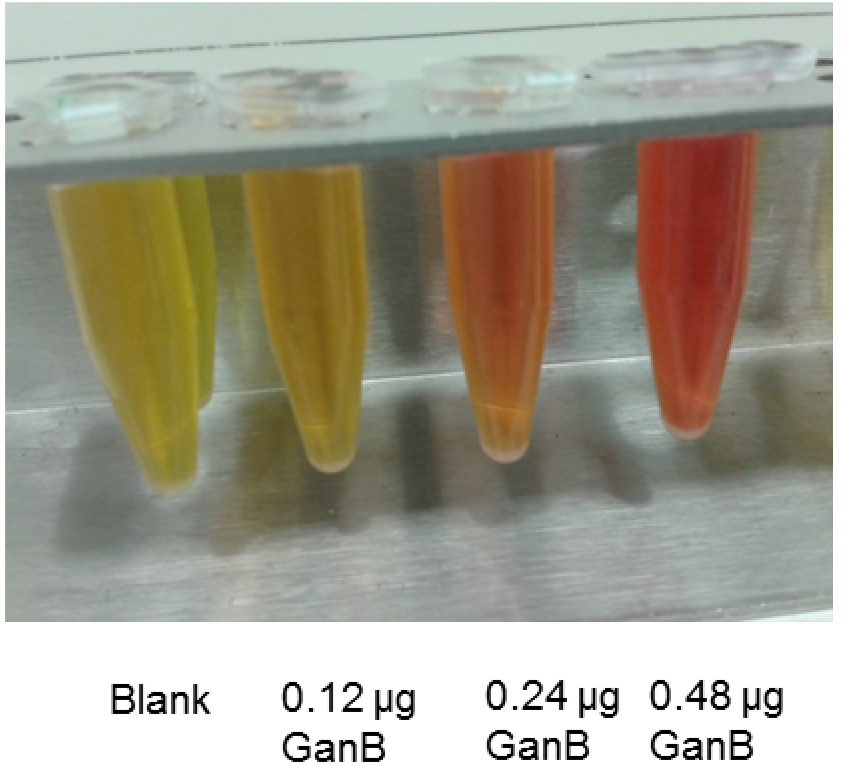
Figure 1. DNS-assay for determination of the reducing power from the reaction of His6-GanB with galactan: Color development of different amounts of His6-GanB after step A5
- Mix appropriate amounts of His6-GanB (maximal 0.5 µg) in 40 µl of 0.1 M potassium phosphate buffer (KPP) pH 6.5 with 40 µl of 2% lupin galactan in an Eppendorf tube.
- Colorimetric assay: Activity on the dyed substrate AZCL-galactan (Azurine-crosslinked-potato galactan) by measurement of the water soluble dyed fragments after degradation of the insoluble AZCL-galactan.
- Delude appropriate amounts of His6-GanB (e.g., 2 µl of 0.6 mg/ml; 2 µl of 0.12 mg/ml; 2 µl of 0.06 mg/ml) in 200 µl of 0.1 M KPP pH 6.5 and mix with 50 µl of a well mixed suspension of 1% AZCL-galactan in an Eppendorf tube.
- Incubate the Eppendorf tube in a thermomixer at 37 °C for 15 min with agitation at 300 rpm.
- Stop the reaction by adding of 250 µl of 0.4 M sodium borate.
- Centrifuge at 13,000 x g for 5 min.
- Take 100 µl of the blue colored supernatant and dilute with 900 µl of H2O.
- Measure the absorbance at 595 nm (for 1.2 µg His6-GanB an OD595 of 0.43 was determined).
As the blank the reaction mixture ingredients without enzyme was used.
Figure 2 shows the development of blue color in step B3 (before centrifugation) from the degradation of the unsoluble AZCL-galactan suspension with different amounts of GanB-enzyme. Tube 1 represents the blank.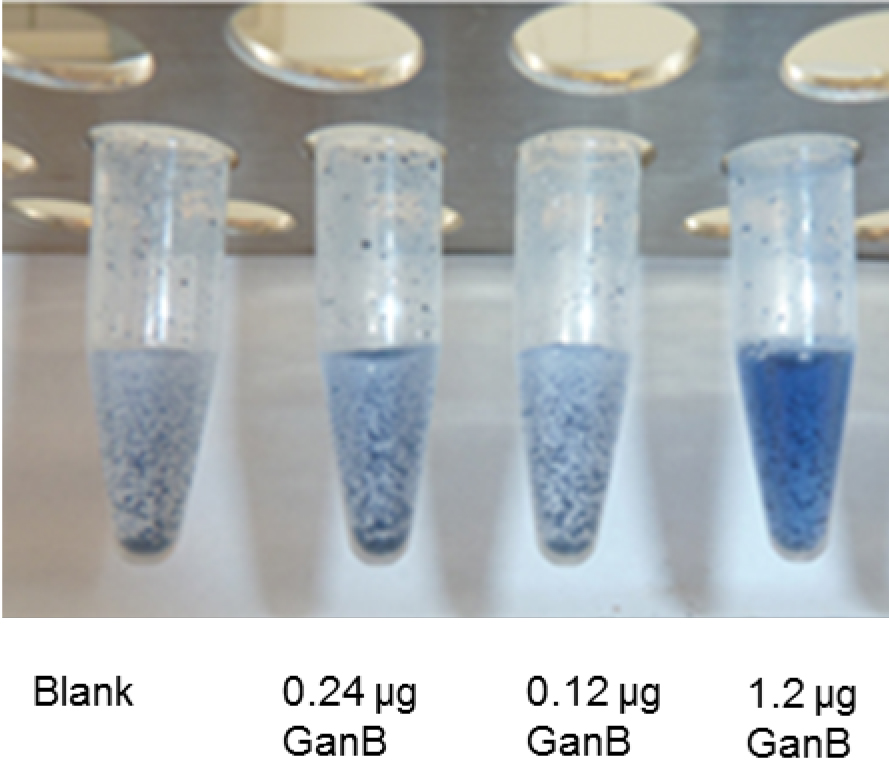
Figure 2. AZCL-galactan assay: Development of the blue color of different amounts of His6-GanB after step B3
Note: The dyed substrate AZCL-galactan is used for semi-quantitatively activity determination as molar extinction coefficient ε595 of the blue dye was unknown.
- Delude appropriate amounts of His6-GanB (e.g., 2 µl of 0.6 mg/ml; 2 µl of 0.12 mg/ml; 2 µl of 0.06 mg/ml) in 200 µl of 0.1 M KPP pH 6.5 and mix with 50 µl of a well mixed suspension of 1% AZCL-galactan in an Eppendorf tube.
- Enzymatic reaction for analysis of the generated galacto-oligosaccharides
- Mix appropriate amounts of His6-GanB (e.g., 1 µg) in 40 µl 0.1 M KPP pH 6.5 with 40 µl of 2% lupin-galactan in an Eppendorf tube and incubate the reaction at 37 °C with agitation at 300 rpm.
- Withdraw 10 µl of the reaction mixture at different times of reaction and put it in a new Eppendorf tube
- Stop the reaction by heating for 5 min at 95 °C.
- Centrifuge at 13,000 x g for 5 min.
- Use aliquots of the heat-denatured enzymatic reaction for analyzing the products by TLC (Procedure D step 1) or HPLC (Procedure E step 1).
- Mix appropriate amounts of His6-GanB (e.g., 1 µg) in 40 µl 0.1 M KPP pH 6.5 with 40 µl of 2% lupin-galactan in an Eppendorf tube and incubate the reaction at 37 °C with agitation at 300 rpm.
- TLC-analysis
- Spot 4 µl of the heat-denatured enzymatic reaction, withdrawn at different reaction times from Procedure C, on a TLC-plate (Silica Gel 60 F254).
- As a control spot 2 µl of a solution of galactose (10 mg/ml) and 2 µl of 1% lupin-galactan on the plate.
- Separate the products with the mobile phase acetone-H2O (87:13) in a thin-layer-chamber till the migration front reach the top.
- Dry the TLC-plate with a blow-dryer approximately 5 min.
- Soak the TLC-plate in the developing reagent.
- Back the plates in an oven at 80 °C till blue spots are visible.
- Figure 3 shows the TLC-chromatogram after separation and visualization of the reaction products generated from lupin galactan degradation of GanB after different reaction times.

Figure 3. TLC-Chromatogram of the degradation products of lupin-galactan by His6-GanB (60 ng) withdrawn at reaction times of 30 min, 1, 2, 4 and 8 h from Procedure C. The reaction products were separated with the mobile phase acetone-H2O (87:13) and visualized by the developing reagent.
- Spot 4 µl of the heat-denatured enzymatic reaction, withdrawn at different reaction times from Procedure C, on a TLC-plate (Silica Gel 60 F254).
- HPAEC-analysis
- Dilute samples of the enzymatic reaction from Procedure C 1:10 with H2O.
- Load 10 μl on the CarboPac PA1 column by injection in the HPLC apparatus and separate the products with an isocratic mobile phase of 0.22 N NaOH/0.02 M sodium acetate on a HPLC-System at a flow rate of 0.75 ml/min.
- Detect the eluted carbohydrate by pulsed amperometry with an analytical cell (5040, ESA Coulochem II, Bischoff).
- Individual sugars like galactose and β-1,4 galactobiose were identified by comparing retention times with those of standards.
- A typical HPLC-profile is given in Figure 4.
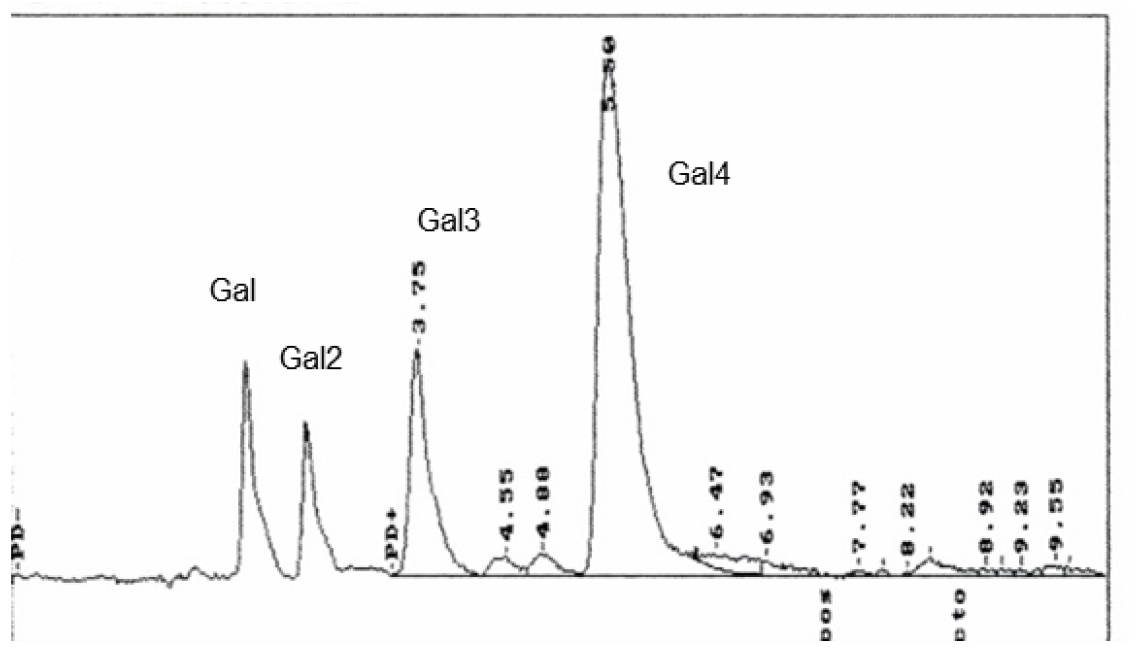
Figure 4. HPLC-chromatogram of the degradation products of lupin-galactan by purified His6-GanB of the reaction of 1 µg His6-GanB in 40 µl 0.1 M KPP pH 6.5 and 40 µl 2% Galactan after incubation at 37 °C for 30 min. The identity of the individual sugars were galactose (Gal); β1,4-galactobiose (Gal2), Galactotriose (Gal3) and Galactotetraose (Gal4).
- Dilute samples of the enzymatic reaction from Procedure C 1:10 with H2O.
Data analysis
- During the enzymatic assays each value was determined in triplicates.
- The reducing power during the galactan degradation by GanB described in Procedure A was determined using the DNS assay and quantified according to a galactose standard curve given in Figure 5. 1 U of galactanase was defined as the release of 1 µmol galactose per minute and is calculated from the equation:
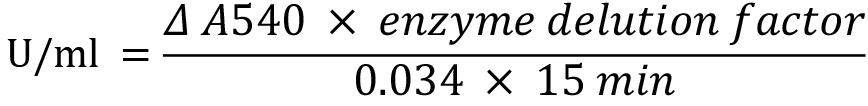

Figure 5. Standard curve of galactose determined by DNS-assay. The A540 nm values were obtained by mixing of 80 µl of a solution of 5 to 20 mM galactose with 300 µl DNS-reagent and proceeded according to Procedure A step 4.
Recipes
- Purified recombinant His6-tagged GanB
0.6 mg/ml in 0.1 M potassium phosphate buffer pH 6.5
Note: The protein quantity was determined by the method of Bradford (Bradford, 1976) using bovine serum albumin as standard. - Lupin-galactan
Dissolve 2% lupin-galactan in Milli-Q pure H2O - DNS-reagent
Mix 1.76 g dinitrosalicylic acid, 50 g potassium-sodium-tartrate, 1.26 g sodium sulfite, 6 ml 50% NaOH with 183 ml Milli-Q pure H2O - AZCL-galactan
Make a suspension of 1% AZCL-galactan in Milli-Q pure H2O and mix thoroughly before using - TLC developing reagent
Mix 10.5 g ammonium heptamolybtate, 0.5 g cerium sulfate, 15 ml concentrated H2SO4 in 245 ml Milli-Q pure H2O - Potassium phosphate buffer (KPP), 0.1 M
Mix the solutions of 0.1 M KH2PO4 and 0.1 M K2HPO4 till the pH reached 6.5 - 0.4 M sodium borate solution
Dissolve 152 g sodium borate in 1 L Milli-Q pure H2O and adjust the pH to 9.8 - 0.3 M sodium acetate, pH 5.2
Dissolve 40.8 g sodium acetate in 1 L Milli-Q pure H2O and adjust the pH to 5.2 with acetic acid - TLC mobile phase solution
Mix 87 ml acetone with 13 ml Milli-Q pure H2O - HPAEC isocratic mobile phase
Mix 11 ml 50% sodium hydroxide, 63.7 ml 0.3 M sodium acetate pH 5.2 with 890.3 ml Milli-Q pure H2O
Acknowledgments
This protocol was first used in Watzlawick et al. (2016).
References
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3): 426-428.
- Watzlawick, H., Morabbi Heravi, K. and Altenbuchner, J. (2016). Role of the ganSPQAB operon in degradation of galactan by Bacillus subtilis. J Bacteriol 198(20): 2887-2896.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Watzlawick, H. (2017). Measurement of the Galactanase Activity of the GanB Galactanase Protein from Bacillus subtilis. Bio-protocol 7(7): e2206. DOI: 10.21769/BioProtoc.2206.
Category
Microbiology > Microbial biochemistry > Protein
Microbiology > Microbial biochemistry > Carbohydrate
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









