- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Xylem Sap Extraction Method from Hop Plants
(*contributed equally to this work) Published: Vol 7, Iss 6, Mar 20, 2017 DOI: 10.21769/BioProtoc.2172 Views: 12585
Reviewed by: Zhaohui LiuJoëlle SchlapferTimo Lehti

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Reliable In Planta Inoculation and Antifungal Screening Protocol for Rhizoctonia solani-Induced Sheath Blight in Rice
Alinaj Yasin [...] Palash Deb Nath
Nov 5, 2025 1582 Views

Quantitative Analysis of the Arabidopsis Leaf Secretory Proteome via TMT-Based Mass Spectrometry
Sakharam Waghmare [...] Rucha Karnik
Nov 20, 2025 2025 Views

Reproducible Emu-Based Workflow for High-Fidelity Soil and Plant Microbiome Profiling on HPC Clusters
Henrique M. Dias [...] Christopher Graham
Jan 20, 2026 441 Views
Abstract
Verticillium wilt is one of the most important diseases on hop that significantly influence continuation of production on affected areas. It is caused by the soil borne vascular pathogen Verticillium nonalfalfae, which infects plants through the roots and then advances through the vascular (xylem) system. During infection, V. nonalfalfae secretes many different virulence factors. Xylem sap of infected plants is therefore a rich source for investigating the molecules that are involved in molecular interactions of Verticillium – hop plants. This protocol provides instructions on how to infect hop plants with V. nonalfalfae artificially and how to obtain xylem sap from hop plants.
Keywords: Verticillium nonalfalfaeBackground
Extraction of xylem sap from plants is mostly used for studies of xylem sap proteome and various methods have been used to extract sap from plant xylem tissues. Buhtz et al. (2004) used hand-held pipettes to collect xylem sap from cut plant stems for comparing xylem proteomes in different plants (broccoli, oilseed rape, pumpkin and cucumber). The same method was used for collecting xylem sap from Brassica napus (Kehr et al., 2005), Brassica oleracea (Ligat et al., 2011) and soybean (Subramanian et al., 2009). Alvarez et al. (2006) extracted the maize xylem sap proteome using ‘root pressure’, as described by Goodger et al. (2005). Dafoe and Constabel (2009) used a Tygon tube, which was fitted over the wood, to collect xylem sap from hybrid poplar and no additional pressure was applied. Information on the protein content of xylem sap is also available for apple, pear and peach (Biles and Abeles, 1991).
Because vascular plant fungal pathogens spread inside host plants through their xylem, this fluid is the most appropriate medium to search for in planta secreted virulence factors from the fungal pathogen. Rep et al. (2002) used a simple xylem sap extraction method by Satoh et al. (1992) whereby cut stems of Fusarium oxysporum f. sp. lycopersici-infected tomato were placed in a horizontal position and sap was dripped from the cut surface. A Scholander pressure chamber (Scholander et al., 1965) was used to obtain xylem sap from Verticillium longisporum-infected oilseed rape (Floerl et al., 2008).
It is hard to draw any conclusion as to which method works best for what kind of plant since no comparative studies have been performed on different methods on the same plant species. It appears that plant species with soft and juicy stems need no pressure added to obtain xylem sap and simple methods are efficient for xylem sap extraction. This may be related to the root pressure, which causes xylem sap to rise through a plant stem from the roots towards the leaves due to osmosis in the roots (Taiz and Zeiger, 2010). However, some species never generate any root pressure (Kramer and Boyer, 1995) so some external force (e.g., a pressure chamber) must be provided in order to extract xylem sap from such plants. So far, there is no specific extraction method available for collecting xylem sap from hop plant infected with Verticillium nonalfalfae. Hop plants have woody roots and exposure of roots to pressure causes no harm to root tissue and no contamination (e.g., with cytosol liquids, cell membranes and other parts). We therefore used a Scholander pressure chamber on hops. This is the first protocol for sampling xylem sap from hop plants.
Materials and Reagents
- Miracloth (EMD Millipore, catalog number: 475855-1R )
- Filter paper (LLG, catalog number: 9.045 840 )
- Tape
- Plastic bag
- Sterile pipet tips
- 2 ml microtubes (BRAND, catalog number: 780550 )
- Silicone tubing, plastic tubing, glass tubing, etc.
- Petri dish (Golias, catalog number: PE01K )
- Host plants (hop Humulus lupulus, susceptible cultivar ‘Celeia’ and resistant cultivar ‘Wye Target’)
- Fungal conidia (Verticillium nonalfalfae; lethal pathotype PV1 [isolate T2]) (Radisek et al., 2006)
- Fertilizer YaraKristalon yellow NPK 13-40-13 + ME [ME - trace elements: B - 0.025%; Cu * - 0.01%; Fe * - 0.07%; Mn * - 0.04%; Mo - 0.004%; Zn * - 0.025%; * - Chelate base] (Yara International ASA)
- Fertilizer YaraKristalon special NPK 18-18-18 + ME [ME - trace elements: B - 0.025%; Cu * - 0.01%; Fe * - 0.07%; Mn * - 0.04%; Mo - 0.004%; Zn * - 0.025%; * - Chelate base] (Yara International ASA)
- Growing medium:
- For fungus inoculum preparation: liquid GFM – general fungal medium (Kayser, 1992) (see Recipes)
- For plants: sterile soil substrate for growing plants
- For fungus re-isolation: potato dextrose agar + antibiotics (streptomycin sulphate, neomycin, chloramphenicol; each 100 mg/ml) = PDA + A plates (see Recipes)
- Streptomycin sulphate (Duchefa Biochemie, catalog number: S0148 )
- Chloramphenicol (Sigma-Aldrich, catalog number: C0378 )
- Sterile distilled water (IDT, catalog number: 231-791-5 )
- Protease inhibitor cocktail (Sigma-Aldrich, catalog number: P8340 )
- 96% ethanol
- Peptone (Sigma-Aldrich, catalog number: 73049-73-7 )
- Yeast extract (AMRESCO, catalog number: J850 )
- Glucose (Kemika, catalog number: 07051 )
- Potassium nitrate (KNO3) (EMD Millipore, catalog number: 105063 )
- Potato dextrose agar (Biolife, catalog number: 4019352 )
- Neomycin (Duchefa Biochemie, catalog number: M0135 )
Equipment
- Plastic pots (1 L, 2 L)
- 500 ml Erlenmeyer flask (BRAND, catalog number: 92824 )
- Rotary shaker (Infrost, catalog number: 29313 )
- 2 L plastic cup (BRAND, catalog number: 87822 )
- Growth chamber (Kambič Laboratory Equipment, model: RK-13300 )
- Fluorescent grow lamp (Idealo, model: Osram Fluora L 58 W/77 )
- Thoma counting chamber (BRAND, Wertheim, Germany)
- Wooden sticks for hop’s bine support
- Scalpel and tweezers
- Light microscope (Nikon Instruments)
- Scholander pressure chamber (Soilmoisture Equipment, model: 3005 )
- Polystyrene box with ice
Procedure
- Preparation of host plants
- Use one-year-old potted hop plants multiplied from softwood cuttings of the susceptible cultivar ‘Celeia’ and resistant ‘Wye Target’ and grown in 1 L plastic pots. It is important that plants have strong, thick stems, the diameter of which should be more than 5 mm (Figure 1). Use plants are produced by a professional grower. Grow plants in a greenhouse on mist benches. Water the plants every 2 days and fertilize once per week using foliar fertilizers that contain macro and micro elements (e.g., 0.1% solution of YaraKristalon yellow NPK 13-40-13 + ME). To maintain the appropriate health status, spray the plants once per week against pests and foliar diseases using protective plant protection products.

Figure 1. Example of stems of hop plants, cultivar Celeia. Left – strong, thick stem. Right – weak, thin stem.
- Use one-year-old potted hop plants multiplied from softwood cuttings of the susceptible cultivar ‘Celeia’ and resistant ‘Wye Target’ and grown in 1 L plastic pots. It is important that plants have strong, thick stems, the diameter of which should be more than 5 mm (Figure 1). Use plants are produced by a professional grower. Grow plants in a greenhouse on mist benches. Water the plants every 2 days and fertilize once per week using foliar fertilizers that contain macro and micro elements (e.g., 0.1% solution of YaraKristalon yellow NPK 13-40-13 + ME). To maintain the appropriate health status, spray the plants once per week against pests and foliar diseases using protective plant protection products.
- Inoculum preparation
- Place a small piece of fungal mycelium (of the size of a 2 mm ‘ball’) from stock PDA plates in each of four 500 ml Erlenmeyer flasks filled with 250 ml of liquid GFM, supplemented with 100 mg/ml streptomycin sulphate and 100 mg/ml chloramphenicol.
- Incubate for approximately 5 days at 100 rpm on a rotary shaker at room temperature and in the dark (place the shaker in a dark room) (Figure 2).

Figure 2. A picture of fungal culture grown after 5 days. White fungal mycelium in a sphere shape can be seen. Medium with fungal isolates that produce a lot of spores is duller. - Prepare the conidia inoculum (spore size is 5 to 7 μm) by filtration of mycelia and the spore through miracloth (typical pore size is 22-25 µm): place a non-autoclaved piece of Miracloth (20 x 20 cm) on top of a 2 L plastic cup, hand held, and filter each flask separately, one by one. Wash spores and suspend in sterile distilled water. If the concentration is low, all the filtration material is needed.
- Use a Thoma counting chamber to determine the spore concentration. Determine the spore concentration four times for one filtrated suspension and calculate an average.
- Adjust the spore concentration to 5 x 106 conidia/ml with sterile distilled water.
- The final volume of inoculum with a concentration of 5 x 106 conidia/ml for artificial infection should be 1 L. Use a maximum 12 plants per one volume of inoculum.
- Place a small piece of fungal mycelium (of the size of a 2 mm ‘ball’) from stock PDA plates in each of four 500 ml Erlenmeyer flasks filled with 250 ml of liquid GFM, supplemented with 100 mg/ml streptomycin sulphate and 100 mg/ml chloramphenicol.
- Artificial infection
- Uproot one-year-old hop plants that have been grown in 1 L plastic pots. Remove as much soil substrate as possible from the roots by hand.
- Rinse the roots in sterile water (just gently dip for a few seconds).
- Then dip the roots for 10 min in 1 L of inoculum that has been poured into a 2 L plastic cup.
- Inoculate a minimum of 30 plants per treatment.
- Dispose of the remaining fungal inoculum after autoclaving.
- Treat the control plants similarly, but dip their roots in sterile distilled water.
- Pot the plants in new pots of size 2 L.
- Grow the plants as a single bine in a growth chamber (RK-13300, Kambič) under a 12-h photoperiod of fluorescent light (L 58 W/77; Fluora, Osram) at a temperature of 22 °C and relative humidity of 65% during the light period and 20 °C and 70% during the dark period. During growth in the chamber, water the plants twice a week and fertilize once per week using foliar fertilizer containing a higher quantity of nitrogen (e.g., 0.2% solution of YaraKristalon special NPK 18-18-18 + ME).
- Uproot one-year-old hop plants that have been grown in 1 L plastic pots. Remove as much soil substrate as possible from the roots by hand.
- Xylem sap extraction
- Sample the plants 30 days post inoculation (± 3 days).
- Uproot the hop plants. Carefully remove the soil from roots but do not wash them in water.
- Cut the plants 5-10 cm above ground with a scalpel or blade (Figure 3). Use the lower part of the plant (root side with 5-10 cm of stem) for xylem sap extraction and discard the upper part of the plant (containing the rest of the stem and leaves). Wash the cut surface with sterile distilled water (drip a few drops of water onto the surface using a 100 µl pipette and soak the water up with filter paper).

Figure 3. Plants are cut 5-10 cm above ground with a scalpel or blade - Place the root in a plastic bag (Figure 4), sealed with tape against the base of the stem. The plastic bag does not have to be sealed very well because it is used only to protect the Scholander pressure cylinder from getting dirty. Insert the stem through the metal cover of the Scholander pressure cylinder (Figure 5).

Figure 4. The root is placed in a plastic bag
Figure 5. Metal cover of Scholander pressure cylinder with cut plant stem. The plant stem is placed through the upper part of the Scholander pressure cylinder. - Place the root side in the Scholander pressure chamber (Scholander et al., 1965) and make an installation from sterile pipette tips for leading the xylem sap to a microtube, which is placed on ice (Figure 6). Any other installation (e.g., silicone tubing, plastic tubing, glass tubing, etc.) can be used for leading the xylem sap to the microtube.
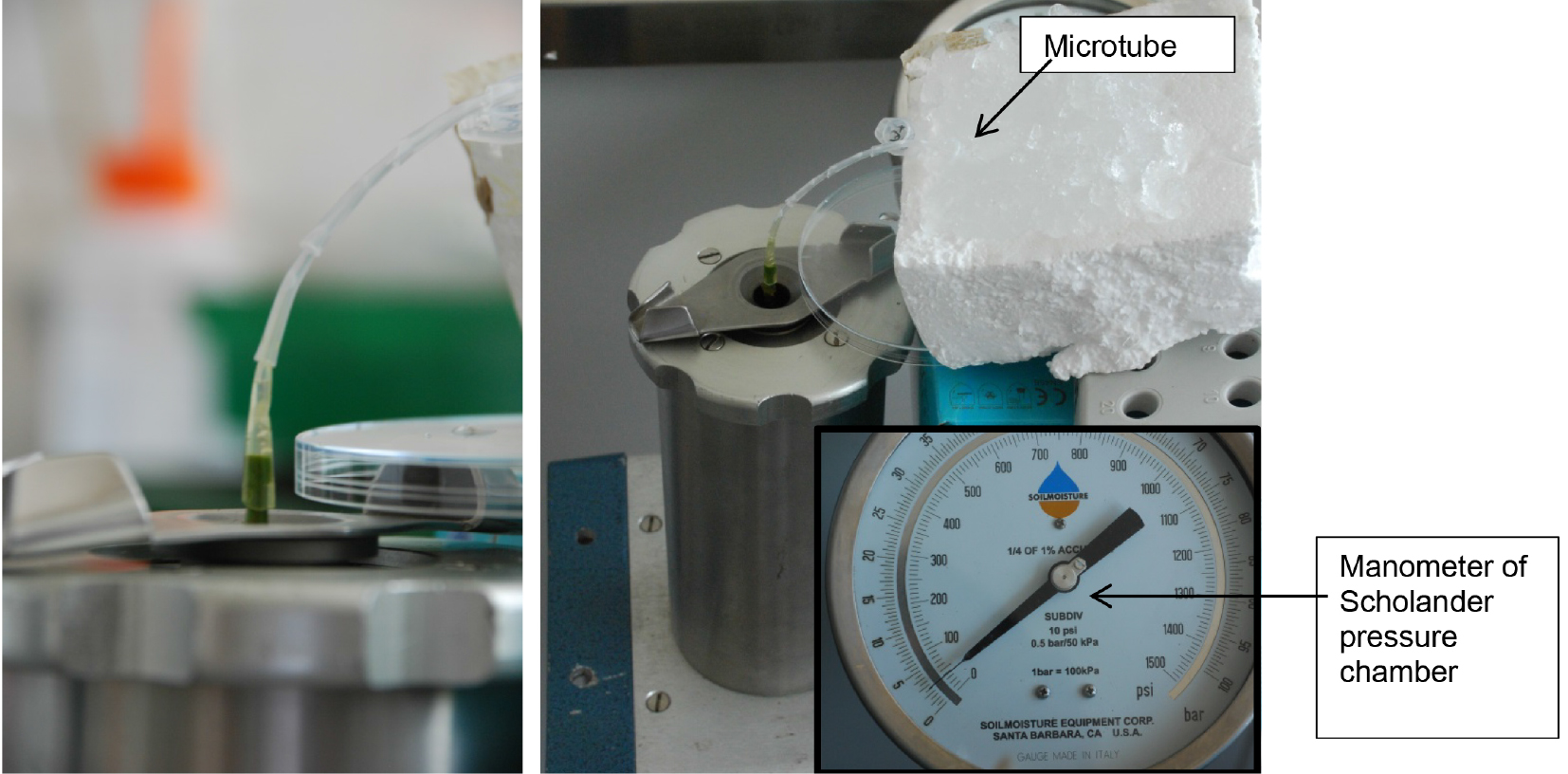
Figure 6. Xylem sap extraction with Scholander pressure chamber. Left: Installation from pipette tips for leading xylem sap to microtube; right: Extraction of xylem sap using Scholander pressure chamber. The microtube is placed on ice, applying a pressure of 2-2.5 bar. - Apply a pressure of 2-2.5 bar to extract xylem fluid.
- Blot the first drops of exuding fluid with filter paper to avoid phloem contamination.
- Then collect the xylem sap (Figure 7) for up to 3 h in microtubes (they must be placed on ice!) containing up to 20 µl 1x protease inhibitor cocktail.
- Store samples of xylem sap at -80 °C until analysis.

Figure 7. The stream of xylem sap. Xylem fluid is seen in the installation from pipette tips. - Use the root side for mycological re-isolation of the pathogen to confirm the presence of the pathogen in the inoculated plants. Perform re-isolation in a laminar. Spray the stems with 96% ethanol and place them very briefly on an open flame in order to sterilise the surface. Harvest the xylem sections from inoculated plants and place them on PDA + A plates. The xylem section is actually the whole inner part of the stem, which should be cut into 1-2 cm long pieces. Place three to six pieces from each plant on PDA + A plates. Fungal outgrowth can be detected after the plates have been incubated for 3-5 days in the dark at room temperature (Figure 8). Examine the emerging mycelium (if any) by light microscopy.
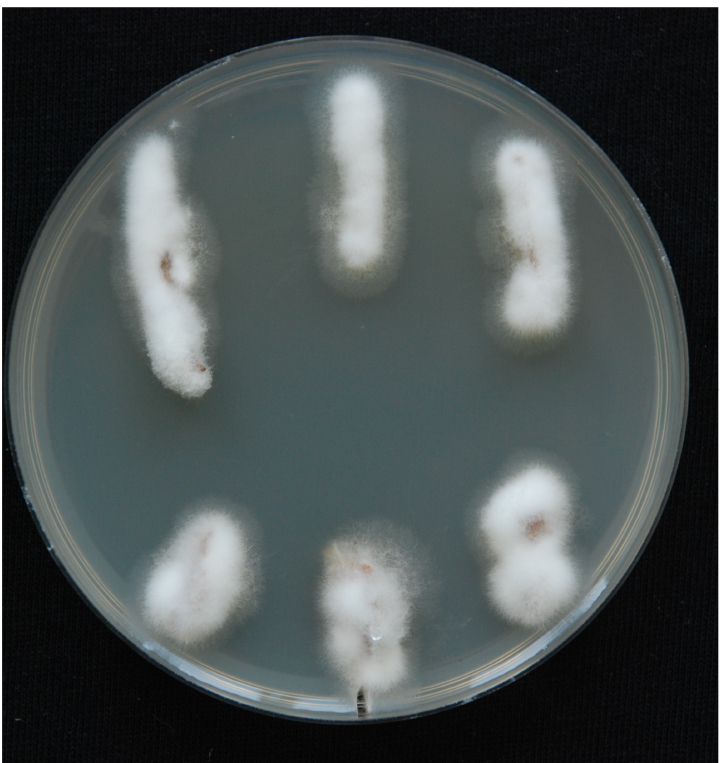
Figure 8. Fungal outgrowth from xylem sections after 3-5 days. White and fluffy mycelium is typical of V. nonalfalfae. Fungal mycelia prefer to grow on the plant material (xylem sections are completely overgrown by mycelium) and not on the PDA medium.
- Sample the plants 30 days post inoculation (± 3 days).
Data analysis
- Up to 2 ml of xylem sap is usually collected from a single plant in a 3 h extraction.
- A very low protein concentration in xylem is expected; samples of 3-10 individual plants (5-10 ml of xylem sap) are therefore pooled into one biological replicate. In our experience, pooled samples have an estimated concentration in the range 5-20 ng/µl.
Notes
- As mentioned in the section ‘Procedure’ (part ‘B. Inoculum preparation’, step 6), a maximum 12 plants per one volume of inoculum (which is 1 L with a concentration of 5 x 106 conidia/ml) should be used. During root dipping of these plants, the inoculum gets diluted. If there are more plants to be inoculated with the same inoculum, the concentration of used inoculum must therefore be recovered to 5 x 106 conidia/ml by adding fresh conidia. It is even better to prepare new inoculum with a concentration of 5 x 106 conidia/ml.
- We recommend that plants are watered one day before xylem sap sampling.
- Regarding xylem sap extraction
- The plants for sap extraction should be cut 5-10 cm above ground. We suggest not cutting higher because we have observed that we obtain a smaller amount of xylem sap with longer stems.
- For every new plant, use a sterile scalpel to cut the stem, new filter paper to blot phloem drops and a new pipette tips installation leading the xylem sap to the microtube.
- When the plant stem is placed through the metal cover of the Scholander pressure chamber, the rubber wrap around stem should not be tightened too much, in order to allow the xylem fluid to run out.
- We recommend collection of xylem sap for a maximum three hours, because prolonging the time does not help to increase the amount of xylem fluid.
- After three hours of collection, remove the installation from pipette tips that lead the xylem sap to the microtube from the cut stem and manually empty by hand-held pipette. Around 100 µl of xylem fluid is recovered with this step.
- The user of this protocol can expect that no xylem fluid will be obtained from some plants, for unknown reasons.
- Emerging mycelium on re-isolation plates must be subjected to morphological analysis using light microscopy in order to confirm the presence of V. nonalfalfae. Aerial mycelium of V. nonalfalfae is generally abundant, floccose to pruinose, hyphae are smooth-walled and 1.5-3 µm wide. Conidiophores are erect or slanted, generally determinate, branched or unbranched, formed disjointedly throughout the colonies and hyaline.
Recipes
- GFM
2 g peptone
2 g yeast extract
20 g glucose
1 g KNO3
Mix in 1,000 ml distilled water and autoclave for 20 min - PDA with 300 mg/ml antibiotics
- Add 35 g of potato dextrose agar in 1,000 ml distilled water
- Shake and mix the powder and autoclave it for 20 min
- Cool down the PDA broth to 55 °C
- Add 1 ml of 100 mg/ml streptomycin sulphate, 1 ml of 100 mg/ml neomycin and 1 ml of 100 mg/ml chloramphenicol and mix well
- Pour in 90 x 15 mm Petri dishes
Acknowledgments
This protocol is adapted from a previously published paper, Flajsman et al., 2016. We acknowledge the Slovenian Research Agency, research programs P4-0077, for funds. We thank Prof. Dr. Dominik Vodnik from the Chair of Applied Botany of the Biotechnical Faculty for the use of their Scholander pressure chamber.
References
- Alvarez, S., Goodger, J. Q., Marsh, E. L., Chen, S., Asirvatham, V. S. and Schachtman, D. P. (2006). Characterization of the maize xylem sap proteome. J Proteome Res 5(4): 963-972.
- Biles, C. L. and Abeles, F. B. (1991). Xylem sap proteins. Plant Physiol 96(2): 597-601.
- Buhtz, A., Kolasa, A., Arlt, K., Walz, C. and Kehr, J. (2004). Xylem sap protein composition is conserved among different plant species. Planta 219(4): 610-618.
- Dafoe, N. J. and Constabel, C. P. (2009). Proteomic analysis of hybrid poplar xylem sap. Phytochemistry 70(7): 856-863.
- Flajsman, M., Mandelc, S., Radisek, S., Stajner, N., Jakse, J., Kosmelj, K. and Javornik, B. (2016). Identification of novel virulence-associated proteins secreted to xylem by Verticillium nonalfalfae during colonization of hop plants. Mol Plant Microbe Interact 29(5): 362-373.
- Floerl, S., Druebert, C., Majcherczyk, A., Karlovsky, P., Kues, U. and Polle, A. (2008). Defence reactions in the apoplastic proteome of oilseed rape (Brassica napus var. napus) attenuate Verticillium longisporum growth but not disease symptoms. BMC Plant Biol 8: 129.
- Goodger, J. Q., Sharp, R. E., Marsh, E. L. and Schachtman, D. P. (2005). Relationships between xylem sap constituents and leaf conductance of well-watered and water-stressed maize across three xylem sap sampling techniques. J Exp Bot 56(419), 2389-2400.
- Kayser, T. (1992). Protoplasten fusion sowie elektrophoretische Chromosomentrennung und Genkartierung bei filamentösen Pilzen: Penicillium janthinellum, Absidia glauca und Cochliobolus heterostrophus. Dissertation.
- Kehr, J., Buhtz, A. and Giavalisco, P. (2005). Analysis of xylem sap proteins from Brassica napus. BMC Plant Biol 5: 11.
- Kramer, P. J. and Boyer, J. S. (1995). Water relations of plants and soils. Academic Press.
- Ligat, L., Lauber, E., Albenne, C., San Clemente, H., Valot, B., Zivy, M., Pont-Lezica, R., Arlat, M. and Jamet, E. (2011). Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 11(9): 1798-1813.
- Radišek, S., Jakše, J. and Javornik, B. (2006). Genetic variability and virulence among Verticillium albo-atrum isolates from hop. Eur J Plant Pathol 116(4): 301-314.
- Rep, M., Dekker, H. L., Vossen, J. H., de Boer, A. D., Houterman, P. M., Speijer, D., Back, J. W., de Koster, C. G. and Cornelissen, B. J. (2002). Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato. Plant Physiol 130(2): 904-917.
- Satoh, S., Iizuka, C., Kikuchi, A., Nakamura, N. and Fujii, T. (1992). Proteins and carbohydrates in xylem sap from squash root. Plant Cell Physiol 33(7): 841-847.
- Scholander, P. F., Bradstreet, E. D., Hemmingsen, E. A. and Hammel, H. T. (1965). Sap pressure in vascular plants: Negative hydrostatic pressure can be measured in plants. Science 148(3668): 339-346.
- Subramanian, S., Cho, U. H., Keyes, C. and Yu, O. (2009). Distinct changes in soybean xylem sap proteome in response to pathogenic and symbiotic microbe interactions. BMC Plant Biol 9: 119.
- Taiz, L. and Zeiger, E. (2010). Plant physiology 5th Ed. Sinauer Associates.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Flajšman, M., Mandelc, S., Radišek, S. and Javornik, B. (2017). Xylem Sap Extraction Method from Hop Plants. Bio-protocol 7(6): e2172. DOI: 10.21769/BioProtoc.2172.
Category
Plant Science > Plant biochemistry > Protein
Plant Science > Plant immunity > Host-microbe interactions
Microbiology > Microbe-host interactions > Fungus
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link













