- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Liposome Flotation Assays for Phosphoinositide-protein Interaction
Published: Vol 7, Iss 5, Mar 5, 2017 DOI: 10.21769/BioProtoc.2169 Views: 19203
Reviewed by: Andrea PuharDaniel KrausVenkatasalam Shanmugabalaji

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Technique for the Measurement of in vitro Phospholipid Synthesis via Radioactive Labeling
Lucia Rodriguez-Berdini and Gabriel O. Ferrero
Jan 20, 2016 14636 Views

Hyaluronan Isolation from Mouse Mammary Gland
Cornelia Tolg [...] Eva A. Turley
Jun 5, 2018 6089 Views

Enhanced Ribonucleoprotein Immunoprecipitation (RIP) Technique for the Identification of mRNA Species in Ribonucleoprotein Complexes
Saja A. Fakhraldeen [...] Caroline M. Alexander
Oct 5, 2022 2861 Views
Abstract
Phosphoinositides are rare membrane lipids involved in the control of the major cellular functions and signaling pathways. They are able to recruit specific effector proteins to the cytosolic face of plasma membrane and organelles to coordinate a vast variety of signaling and trafficking processes, as well to maintain specific identity of the different subcellular compartments (Di Paolo and De Camilli, 2006; Lemmon, 2003). Therefore, analysis of these effectors’ binding properties and specificity towards different phosphoinositides is crucial for the understanding of their cellular functions. This protocol describes a method to characterize the binding of proteins to different phosphoinositide-containing vesicles.
Keywords: Liposome flotation assayBackground
Several methods exist to analyze protein-phosphoinositide binding and the specificity towards the different members of the phosphoinositide family: lipid overlay, lipid flotation assay and surface plasma resonance (SPR). A Lipid flotation assay consists in the incubation of unilamellar vesicles with the protein of interest, and the subsequent flotation of the vesicles on a sucrose cushion, which will separate vesicle-bound proteins from unbound proteins and vesicles alone. Compared to the other methods a lipid flotation assay is i) technically easier and cheaper than SPR and ii) more specific and closer to physiological conditions, as it mimics the curvature of membranes, in contrast to protein-overlay assays, where lipids are dried on a flat nitrocellulose membrane. This protocol describes the binding of recombinant GST-tagged proteins to unilamellar vesicles of defined size and lipid composition, in particular regarding the specificity towards the different monophosphorylated phosphoinositides (PIP).
Materials and Reagents
- Glass tubes, 12 x 75 mm (DUTSCHER SCIENTIFIC, catalog number: 110011 )
- Tube, thickwall, polycarbonate, 1 ml, 11 x 34 mm (Beckman Coulter, catalog number: 343778 )
- Stericup-GP sterile vacuum filter unit, polyethersulfone, 0.22 μm (EMD Millipore, catalog number: SCGPU10RE )
- Mini Extruder Kit (mini-extruder, 2 Hamilton syringes, 0.1 μm polycarbonate membranes, filter supports, holder/heating block) (Avanti Polar Lipids, catalog number: 610000 )
- 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC) (Avanti Polar Lipids, catalog number: 850457 )
- 1-palmitoyl-3-oleoyl-sn-glycero-2-phosphoethanolamine (PE) (Echelon, catalog number: L-2368 )
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(carboxyfluorescein) (fluoPE) (Avanti Polar Lipids, catalog number: 810332 )
- Phosphatidylinositol 3-phosphate diC16 (PI3P) (Echelon, catalog number: P3016 )
- L-α-phosphatidylinositol-4-phosphate (PI4P) from porcine brain (Avanti Polar Lipids, catalog number: 840045 )
- Phosphatidylinositol 5-phosphate diC16 (PI5P) (Echelon, catalog number: P5016 )
- Liquid nitrogen
- Chloroform/methanol (50/50, v/v)
- PBS (w/o Ca/Mg) (Sigma-Aldrich, catalog number: D8537 )
- Sucrose (Sigma-Aldrich, catalog number: S0389 )
- Anti-GST antibody (clone B-14) (Santa Cruz Biotechnology, catalog number: sc-138 )
- Lipid stocks solutions (see Recipes)
- 60% and 25% sucrose solutions in PBS (see Recipes)
Equipment
- Nitrogen evaporator (N-EVAP, Organomation, catalog number: 11250 )
- Chemical hood
- Standard heated water bath
- Optima TLX ultracentrifuge (Beckman Coulter, model: OptimaTM TLX)
- TLA100.2 rotor (Beckman Coulter, model: TLA100.2 rotor)
Note: This rotor is not available anymore from Beckman Coulter. Rotor TLA120.2 (Beckman Coulter, model: TLA120.2) can be used instead. - Portable UV lamp
- SDS-PAGE running apparatus
- Western blotting apparatus
- Refractometer
Software
- Image analysis software (e.g., ImageJ, NIH)
Procedure
- Prepare the large unilamellar vesicles (LUVs)
- To prepare the lipids stock solutions (see Recipes). When using fluorescently labeled lipids, shield from direct light throughout the procedure.
- Prepare a 2x concentrated lipid mix at 1 mM concentration in a glass tube: PC:PE:fluoPE (70:28:2 mol%) to assess basal binding properties and PC:PE:fluoPE:PIP (64:28:2:6 mol%) to assess specific binding properties of the protein to each PIP.
- Evaporate under nitrogen.
- Resuspend twice with chloroform/methanol (v/v) and evaporate under nitrogen to form a nice lipid cake on the glass tube wall. No clumps should remain at this stage.
- Evaporate under nitrogen for a final 30 min-1 h to eliminate any traces of solvent.
- Resuspend in 37 °C pre-warmed PBS and leave to rehydrate for 1 h at RT.
- Perform 5 freeze/thaw cycles in liquid nitrogen/warm water (40 °C). For optimal results, wait for complete freezing/thawing of the samples for each step. Performing these freeze-thaw cycles will help to break the lipid cake and will ensure a good efficiency of the extruder in the following step, therefore improving the results. We found that 5 cycles was providing the best reliability of the results without being too much time-consuming.
- Mount the mini-extruder according to manufacturer’s instructions using the appropriate membrane size (e.g., 0.1 μm polycarbonate membrane for LUVs formation).
- Extrude the lipid mix through the filter, typically 19 times. Collect and transfer into a fresh glass tube.
- To prepare the lipids stock solutions (see Recipes). When using fluorescently labeled lipids, shield from direct light throughout the procedure.
- LUVs-protein mix
- In a thick-wall polycarbonate tube, mix 50 µl of the 2x LUVs with 50 µl of the 2x GST-recombinant protein to have a final volume of 100 µl and a final concentration 0.5 mM LUVs and 1 mM recombinant protein. See Notes section for adaptability. Include as negative control the purified GST protein alone with the LUVs.
- Incubate for 1 h at RT.
- Add 100 μl of 60% sucrose in PBS to yield a 30% final sucrose concentration. Mix gently to have a homogenous solution but without disrupting the interaction. See Figure 1 for a schematic representation.
- Overlay 250 μl of 25% sucrose and 50 μl of PBS buffer on top without disturbing the layers.
- Centrifuge at 174,206 x g for 1 h at 20 °C (corresponding to 70,000 rpm on a TLA100.2 rotor). In order not to disturb the layers and to get reliable results, it is important to use slower acceleration/deceleration parameters than usual, e.g., acceleration 2/deceleration 2 on an OptimaTM TLX ultracentrifuge.
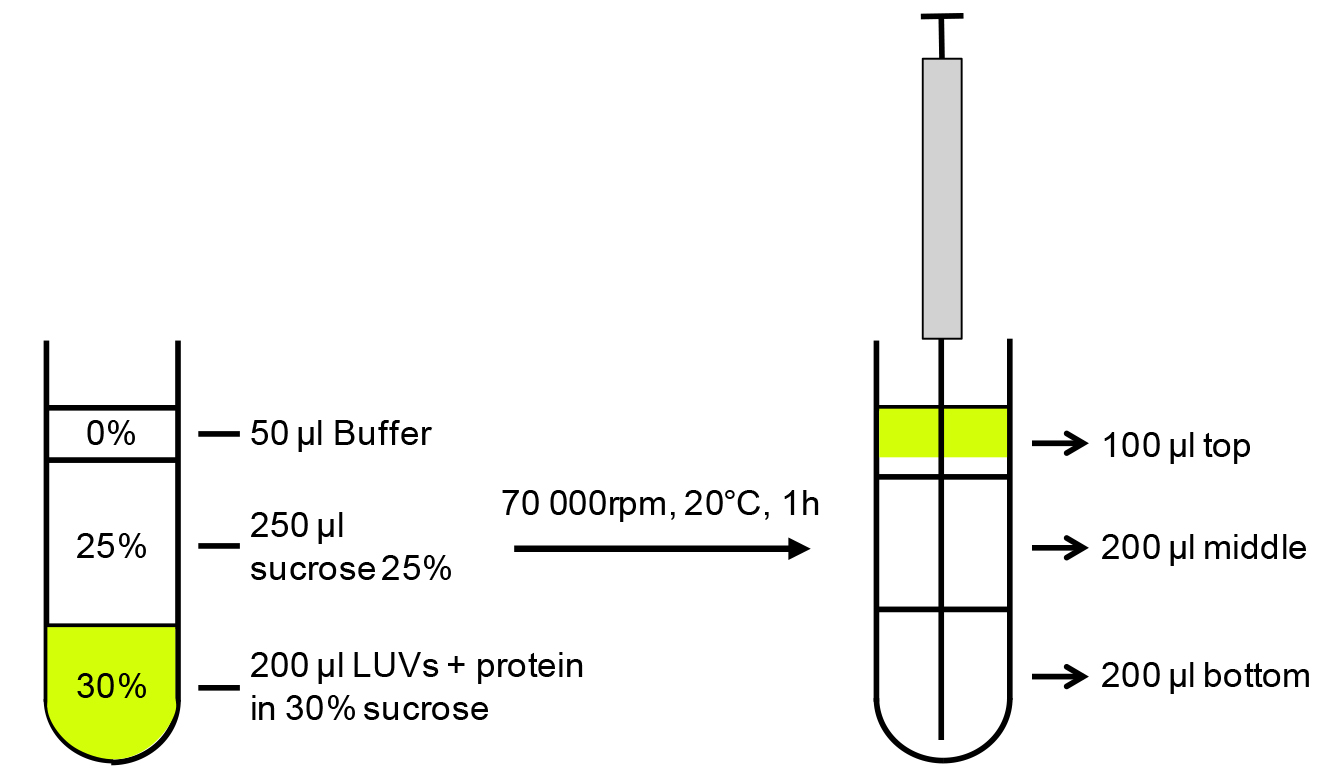
Figure 1. Schematic representation of the liposome binding assay. LUVs and protein mix equilibrated in 30% sucrose are layered with 25% and 0% sucrose, centrifuged to allow LUVs flotation (70 000 rpm [174,206 x g] on a TLA100.2 rotor). Fractions are collected with Hamilton’s syringe from the bottom of the tube.
- In a thick-wall polycarbonate tube, mix 50 µl of the 2x LUVs with 50 µl of the 2x GST-recombinant protein to have a final volume of 100 µl and a final concentration 0.5 mM LUVs and 1 mM recombinant protein. See Notes section for adaptability. Include as negative control the purified GST protein alone with the LUVs.
- Collect the fractions
- As the liposomes will fluoresce owing to the fluo-PE, collect the fractions under a portable UV lamp using a Hamilton syringe, starting from the bottom of the tube (see Figure 1 for schematic representation).
- 200 μl bottom fraction
- 200 μl middle fraction
- 100 μl top fraction containing the LUVs and associated proteins.
- 200 μl bottom fraction
- As the liposomes will fluoresce owing to the fluo-PE, collect the fractions under a portable UV lamp using a Hamilton syringe, starting from the bottom of the tube (see Figure 1 for schematic representation).
Data analysis
Mix 20 µl of the top fraction with sample buffer and resolve by SDS-PAGE. Analyze the amount of recombinant proteins bound to the LUVs by either Coomassie stain or Western blot against the GST moiety or using a specific antibody. Band intensity can be quantified by densitometry using an image analysis software (e.g., ImageJ, NIH) in order to compare binding specificity towards the different PIP. Typically, the GST tag alone does not bind to LUVs, but this has to be checked as a negative control. As shown in Figure 2, we previously used this technique in order to analyze the PIP binding specificity of different structural domains of the endosomal protein TOM1 (Boal et al., 2015). We demonstrated that the VHS domain shows a greater avidity for PI5P, in contrast to the GAT domain (Figure 2B).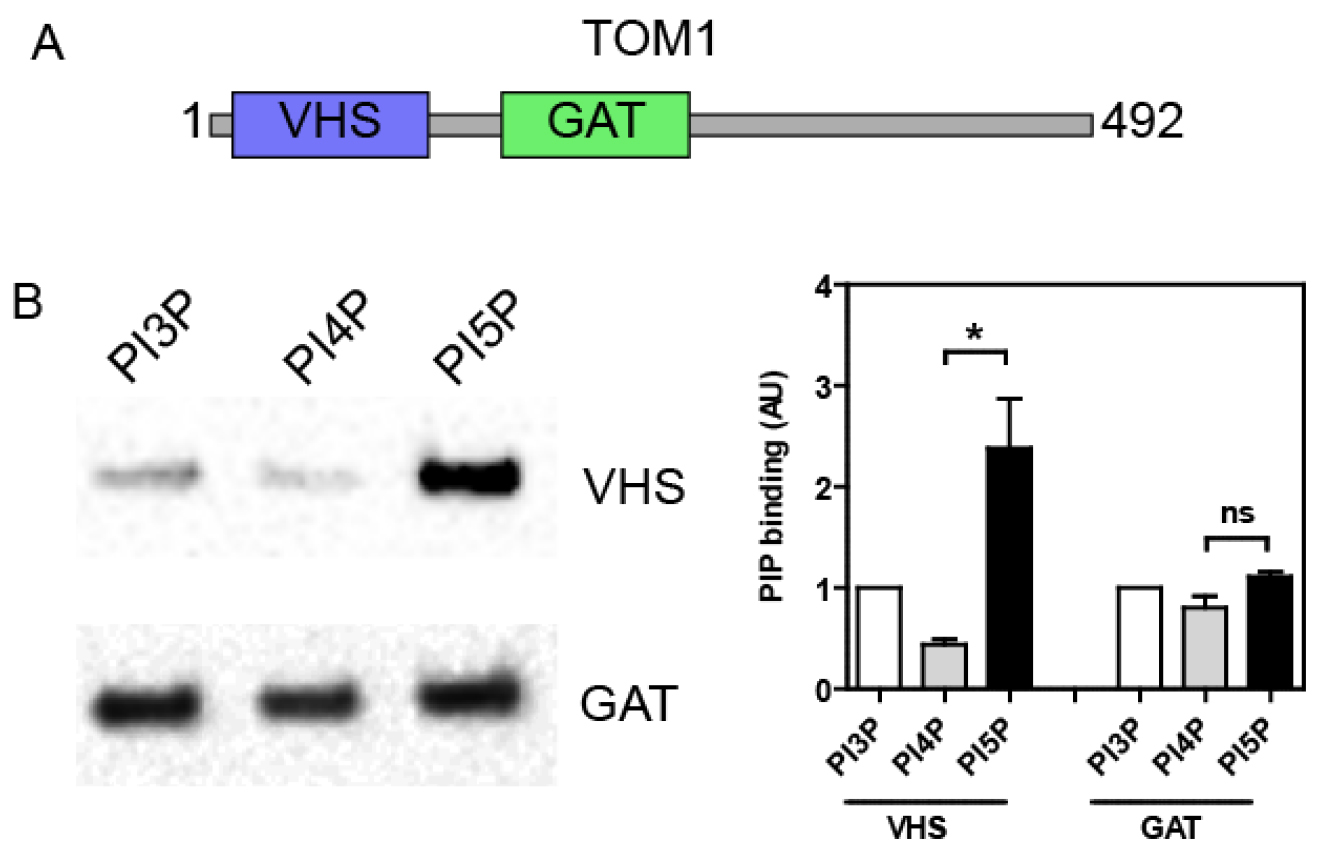
Figure 2. TOM1 VHS but not GAT domain shows specificity towards PI5P binding in a lipid flotation assay. A. TOM1 structural domains; B. N-terminally GST- tagged VHS or GAT domains were incubated with liposomes containing 6% mol of the indicated PIP. After flotation on a sucrose cushion, bound proteins were analyzed by Western blot using an anti-GST antibody (left panel). Right panel shows quantification across 4 independent experiments. Results are presented as mean ± SEM; t-test *P < 0.05; ns, non-significant.
Notes
- Once prepared, LUVs stock solution can be kept in a nitrogen-filled sealed glass tube at 4 °C for a couple of days but one might expect some oxidation and should take care when analyzing the results.
- According to the affinity of the protein towards the LUVs, the final concentration for the recombinant protein can be modified. Similarly, composition of the binding buffer can be adapted, e.g., buffer specificity, calcium-dependence, blocking agent (fatty-acid free BSA).
- Centrifugation step should be performed with gentle acceleration/deceleration in order not to disturb the sucrose layers.
- The use of fluoPE allows a quick and easy visualization of the flotation. When collecting the fractions, one must take care not to disturb the top layer, as that will induce cross-contamination and reduce reproducibility of the results.
- The purity of the recombinant protein is crucial. Indeed, bacterial contaminants could greatly alter the results. Purity must at least be checked by SDS-PAGE followed by Coomassie stain. Protein aggregates must be eliminated by centrifugation at full speed. In case of affinity-purification using the GST-tag, the elution buffer must be exchanged for the binding buffer (e.g., PBS) by dialysis.
Recipes
- Lipid stocks solutions
Prepared in chloroform/methanol at the concentration of 1 mg/ml in glass tubes
Stored at -20 °C
Note: After each use, apply a stream of nitrogen to fill in the glass tube and avoid lipid oxidation but limit evaporation of chloroform/methanol as this will modify the concentration of the stocks. Tighten the cap with Teflon rubber. - 60% and 25% sucrose solutions in PBS
Dissolve the sucrose in PBS (60% eq. 771.9 g/L and 25% eq. 275.9 g/L)
Once fully dissolved, check density with a refractometer and adjust with PBS
Note: As sucrose density fluctuates with temperature, take care to fully equilibrate your buffer. Filtrate with Millipore stericup sterile vacuum filter unit. In order to prevent bacterial contamination, the sucrose stock solutions have to be kept sterile and at 4 °C.
Acknowledgments
This protocol was modified from the work of Bigay and Antonny (2005). This study was supported by grants from Institut National de la Santé et de la Recherche Médicale; Agence Nationale de la Recherche; Fondation pour la Recherche Médicale; The French Muscular Dystrophy Association (AFM); and by the Centre National de la Recherche Scientifique; the Région Midi-Pyrénées and European funds (FEDER, Fonds Européens de Développement Régional). The authors declare no conflict of interest or competing interests.
References
- Bigay, J. and Antonny, B. (2005). Real-time assays for the assembly-disassembly cycle of COP coats on liposomes of defined size. Methods Enzymol 404: 95-107.
- Boal, F., Mansour, R., Gayral, M., Saland, E., Chicanne, G., Xuereb, J. M. and Tronchère, H. (2015). TOM1 is a PI5P effector involved in the regulation of endosomal maturation. J Cell Sci 128(4): 815-827.
- Di Paolo, G. and De Camilli, P. (2006). Phosphoinositides in cell regulation and membrane dynamics. Nature 443(7112): 651-657.
- Lemmon, M. A. (2003). Phosphoinositide recognition domains. Traffic 4(4): 201-213.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tronchere, H. and Boal, F. (2017). Liposome Flotation Assays for Phosphoinositide-protein Interaction. Bio-protocol 7(5): e2169. DOI: 10.21769/BioProtoc.2169.
Category
Cancer Biology > General technique > Biochemical assays > Other compound
Cell Biology > Cell signaling > Intracellular Signaling
Biochemistry > Lipid > Lipid-protein interaction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link