- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Polyethylene Glycol-mediated Transformation of Drechmeria coniospora
Published: Vol 7, Iss 5, Mar 5, 2017 DOI: 10.21769/BioProtoc.2157 Views: 9458
Reviewed by: Arsalan DaudiMichael TschernerAksiniya Asenova

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Phytophthora sojae Transformation Based on the CRISPR/Cas9 System
Jingting Cao [...] Yuanchao Wang
Mar 20, 2022 3490 Views

Simple Growth Complementation Assay in Yeast
Robert Fuhrmeister and Jana Streubel
Aug 5, 2023 2147 Views

Gene Replacement by a Selectable Marker in the Filamentous Fungus Magnaporthe oryzae
Nalleli Garcia [...] Jessie Fernandez
Sep 5, 2023 2146 Views
Abstract
Drechmeria coniospora is a nematophagous fungus and potential biocontrol agent. It belongs to the Ascomycota. It is related to Hirsutella minnesotensis, another nematophagous fungus but, phylogenetically, it is currently closest to the truffle parasite Tolypocladium ophioglossoides. Together with its natural host, Caenorhabditis elegans, it is used to study host-pathogen interactions. Here, we report a polyethylene glycol-mediated transformation method (Turgeon et al., 2010; Ochman et al., 1988) for this fungus. The protocol can be used to generate both knock-in or knock-out strains (Lebrigand et al., 2016).
Keywords: FungusBackground
D. coniospora has been developed as a model pathogen for the study of innate immunity in C. elegans (Lebrigand et al., 2016 and references therein). D. coniospora grows slowly on standard growth media, rendering in vitro study difficult and making it hard to develop transformation methods. We report here a culture method that allows the rapid production of large quantities of D. coniospora, opening the way to its genetic modification. Polyethylene glycol-mediated transformation is probably the simplest method that has been broadly applied to modify fungi. We found that it can be used with D. coniospora, thus providing the first approach to modify its genome deliberately.
Materials and Reagents
- Pasteur pipettes (VWR, catalog number: 6121701 )
- Eppendorf tube 1.5 ml (Sigma-Aldrich, catalog: T9661 )
- Petri dish (Greiner Bio one, catalog number: 633185 )
- 50 ml conical tube (Corning, Falcon®, catalog number: 352070 )
- 15 ml conical tube (Corning, Falcon®, catalog number: 352196 )
- 14 ml polystyrene round bottom tube (Corning, Falcon®, catalog number: 352051 )
- Miracloth (EMD Millipore, catalog number: 475855 )
- Syringe 25 ml (Terumo, catalog number: SS-20ES )
- Acrodisc® syringe filters (Pall, catalog number: 4187 )
- Aluminum foil (Fisher Scientific, catalog number: 01213102 )
- Escherichia coli strain OP50
- Caenorhabditis elegans strain N2
- Drechmeria coniospora recipient strain ATCC 96282
- pLH4237 plasmid DNA, contains a hygromycin B phosphotransferase::GFP chimeric gene driven by D. coniospora β-tubulin promoter (β-tubp::HPH::GFP, Lebrigand et al., 2016)
- Ampicillin sodium salt (Sigma-Aldrich, catalog number: A9518 )
- Gentamicin solution (Sigma-Aldrich, catalog number: G1272 )
- Hygromycin B (Thermo Fisher scientific, GibcoTM, catalog number: 10687010 )
- di-Potassium hydrogen orthophosphate, K2HPO4 (VWR, catalog number: 26931.263 )
- Potassium dihydrogen phosphate, KH2PO4 (VWR, catalog number: 0781-1KG )
- Bacto agar (BD, Bacto, catalog number: 214010 )
- Bacto peptone (BD, Bacto, catalog number: 211677 )
- Sodium chloride, NaCl (VWR, catalog number: 27810.295 )
- Cholesterol (Sigma-Aldrich, catalog number: C3045 )
- Ethanol (VWR, catalog number: 20821.321 )
- Magnesium sulfate, MgSO4 (Sigma-Aldrich, catalog number: M7506 )
- Calcium chloride, CaCl2 (Sigma-Aldrich, catalog number: C1016 )
- Yeast extract (Douchefa Biochemie, catalog number: Y1333 )
- D-sorbitol (Sigma-Aldrich, catalog number: S3889 )
- Aurintricarboxylic acid, ATA (EMD Millipore, catalog number: 189400 )
- Tris/HCl (Sigma-Aldrich, catalog number: T6666 )
- Polyethylene glycol (PEG 3350) (Sigma-Aldrich, catalog number: P4338 )
- Caylase C4 (CAYLA, catalog number: caseC4-5 ) Alternative can be lysing enzymes (Sigma-Aldrich, catalog number: L1412)
Note: The hyperlink for caseC4-5 is not available anymore. Alternative could be lysing enzyme, and we are still testing its efficiency in our lab. - Phosphate buffer (see Recipes)
- Medium (see Recipes)
- NGM-AG
- NGMY
- NGMY liquid
- NGMS
- 15 mM ATA solution (see Recipes)
- 50 mM NaCl solution (see Recipes)
- CaCl2 1700 (see Recipes)
- Lysis buffer (see Recipes)
- STC 1700 (see Recipes)
- PEG 1700 (see Recipes)
Equipment
- Stereomicroscope (Leica Microsystems, model: MZ16 F )
- HERAsafe KS safety cabinet (Thermo Fisher Scientific, Thermo ScientificTM, model: HerasafeTM KS Class II , catalog number: 51022751)
- Centrifuge (Eppendorf, catalog number: 5810 R )
- Funnel
- Spatula
- Cloth-plugged 250 ml glass Erlenmeyer flask
- Rotary shaker for culture flasks (Eppendorf, model: New Brunswick Scientific Innova 4080 )
- Counting chambers (Burker, Tiefe 0.1000 m, 0.0025 mm2)
- Balancer
- Pipetman P1000
- autoclave
Procedure
- Mycelium preparation
- Thaw a frozen stock of fungal mycelia at room temperature and spread on NGM plates (with ampicillin and gentamycin). Culture at 25 °C for 1 week. Using a stereomicroscope, confirm appearance of spores. If no spores are visible, continue culture and check daily.
- Using a bent sterile glass Pasteur pipette, gently rub the fungal colonies to dislodge the spores. Wash the spores from the plates, using 1 ml of 50 mM NaCl (Figures 1A and 1B) and collect into an Eppendorf tube.
- Spread the spores (20-100 µl) on a 9 cm NGM-AG plate (Figure 1C) and allow to dry in a safety cabinet. In parallel, prepare a population of synchronized L4 stage worms (Stiernagle, 2006). Typically, from a single densely populated 9 cm plate, one can obtain 4,000-6,000 worms. Wash the worms from each plate with 5 ml 50 mM NaCl into a 15 ml Falcon tube and centrifuge at 721 x g for 1 min; carefully remove the supernatant, resuspend in the remaining solution and spread half the mix on the 9 cm NGM-AG plate. The worms will be infected by spores. Incubate the resulting plate at 25 °C for 10 days.
- Wash off spores as in step A2. Prepare four 9 cm NGM plates seeded with OP50 (Stiernagle, 2006). Spread the spores collected from one NGM-AG plate onto 4 NGM plates with OP50. If necessary, allow the solution to dry.

Figure 1. Preparation of infection plates. A and B. Washing spores from NGM-AG plate; C. Spreading spore solution on an NGM OP50 plate; D. Spores from NGM-AG plate, photomicrograph using Nomarski optics. - Transfer L4 or young adult N2 worm collected from one 9 cm plate as above (generally 4,000-6,000 worms) onto each spore-OP50 plate, and incubate for 24-30 h.
- Wash the infected worms from each plate with 1.5 ml 50 mM NaCl into a 15 ml Falcon tube and centrifuge at 721 x g for 1 min; carefully remove the supernatant, resuspend and divide the worm pellet between three 9 cm NGMY plates. Gently spread the pellet across the surface of the plate using a bent sterile glass Pasteur pipette. If necessary, allow to dry in a safety cabinet before incubating at 25 °C for 1 day. Most adult worms should be dead by this time (Figure 2). Before dying, they will have produced progeny and most of the worms of the next generation will be alive at this time.
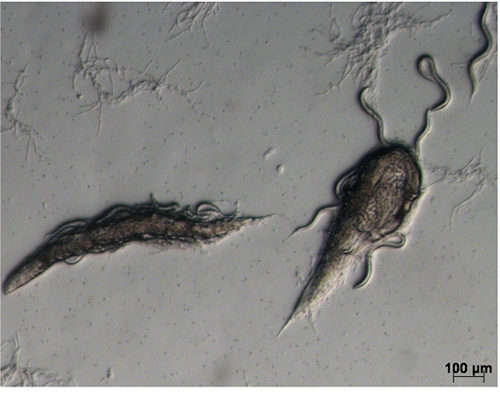
Figure 2. Infected worms on NGMY plate. Two dead adult worms with a number of live larvae. - Add 1-2 ml of 50 mM NaCl to each plate, and using a bent sterile glass Pasteur pipette, rub to resuspend the dead worms. Collect the liquid from 3 plates in a 15 ml Falcon tube; transfer the solution into 250 ml NGMY liquid, culture on a shaking incubator at 170 rpm for 36-40 h.
- Thaw a frozen stock of fungal mycelia at room temperature and spread on NGM plates (with ampicillin and gentamycin). Culture at 25 °C for 1 week. Using a stereomicroscope, confirm appearance of spores. If no spores are visible, continue culture and check daily.
- Protoplasting
- Pour the liquid culture into a funnel lined with Miracloth to filter the mycelia; wash with 100 ml CaCl2 1700 buffer, use a spatula to transfer the mycelia into a 50 ml Falcon tube (Figure 3). Keep the used funnel and Miracloth in a sterile environment for later use.

Figure 3. Collecting mycelium with Miracloth - Weigh the mycelia and resuspend with gentle vortexing in newly prepared lysis buffer; for every 1 g mycelia add 10 ml lysis buffer. If the volume exceeds 30 ml, use a sterile, cloth-plugged Erlenmeyer flask, otherwise a 50 ml Falcon tube.
- Incubate the mycelial solution at 30 °C with 60 rpm for 1-1.5 h. After 1 h, at regular intervals, remove a small aliquot and check the solution under a microscope. Mycelial fragments are quite resistant to hygromycin selection and can give rise to colonies; avoid fragmentation as much as possible. If short mycelial fragments appear (Figure 4), stop the incubation by moving to the next step.
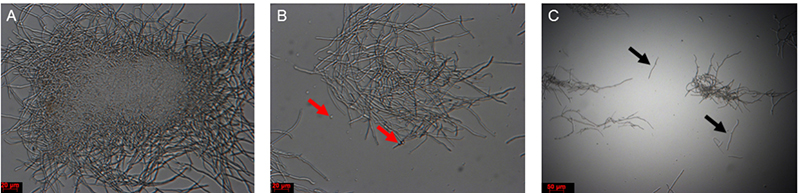
Figure 4. Mycelium solution before and after lysis buffer digestion. A. Colony before treatment; B. Colony after 1 h of treatment, with protoplasts (red arrows); C. Mycelial fragments after treatment (black arrows). Scale bars = 20 μm in A and B; Scale bar = 50 μm in C. - Place the funnel together with the Miracloth used in protoplasting step 1 onto a 50 ml Falcon tube. Filter the mycelia and protoplasts in lysis buffer, retain the filtrate.
- Add an equal volume of ice-cold STC buffer to the protoplast solution, mix gently and leave on ice for 10 min.
- Centrifuge the mixture at 4 °C, 1,127 x g for 15 min.
- Carefully discard the supernatant; firstly, gently resuspend the pellet with 1 ml ice-cold STC buffer by pipetting, then add another 24 ml of cold STC buffer to the protoplast; centrifuge the mixture at 4 °C, 1,623 x g for 10 min.
- Repeat step B7 one more time.
- Discard the supernatant, add 300 µl ice-cold STC buffer to resuspend the protoplasts; remove a small aliquot and count the protoplasts using a haemocytometer (Figure 5). Dilute the protoplasts to a final concentration of 108 protoplasts/ml using ice-cold STC buffer. Then, stock the solution on ice or at 4 °C; it can be used for transformation for a maximum of 7 days.
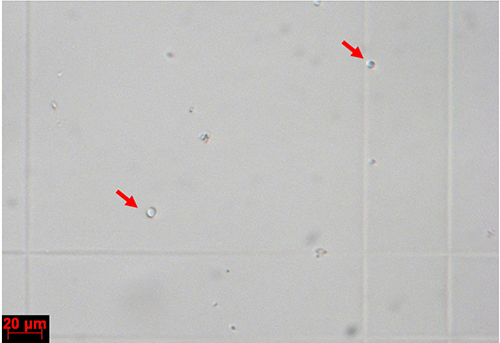
Figure 5. Protoplasts on a haemocytometer. Individual protoplasts are indicated with red arrows.
- Pour the liquid culture into a funnel lined with Miracloth to filter the mycelia; wash with 100 ml CaCl2 1700 buffer, use a spatula to transfer the mycelia into a 50 ml Falcon tube (Figure 3). Keep the used funnel and Miracloth in a sterile environment for later use.
- Transformation
- Add 1 µl ATA onto the wall of a round-bottom 14 ml Falcon tube, mix with 10 µg or more of plasmid DNA (in a volume of around 10 µl), then mix with 150 µl of protoplast solution by gently pipetting; incubate the mixture for 40 min at room temperature.
- Using a Pipetman P1000, add 6 drops of PEG1700 buffer, mix by gently shaking; add another 850 µl PEG buffer, gently shake again, incubate for another 20 min.
- Add 10 ml of room temperature STC buffer; centrifuge for 10 min at 1,623 x g at room temperature.
- Discard the supernatant carefully; resuspend the protoplasts with 300 µl STC at room temperature.
- Add 150 µl of protoplast solution to a 9 cm plate (see Note 2), spread gently with sterile bent glass Pasteur pipette; culture at 25 °C for 3 weeks.
- Check any colonies for successful transformation, by PCR or under a fluorescent stereomicroscope when using a transformation construct that includes a fluorescent reporter gene.
- Add 1 µl ATA onto the wall of a round-bottom 14 ml Falcon tube, mix with 10 µg or more of plasmid DNA (in a volume of around 10 µl), then mix with 150 µl of protoplast solution by gently pipetting; incubate the mixture for 40 min at room temperature.
Notes
- Unless specified, temperature is always at 25 °C.
- Except OP50 NGM plates, all the other plates are with ampicillin and gentamicin. When transformation constructs include a hygromycin-resistance cassette, NGMS plates are supplemented with 25 µg/ml hygromycin.
- Autoclave the Miracloth (folded in half twice), in an aluminum foil bag with a cut corner, so that condensation does not accumulate and the cloth is dry after autoclaving (Figure 6).

Figure 6. Miracloth preparation
Recipes
- 1 M phosphate buffer, pH 6.0
108.3 g KH2PO4
35.6 g K2HPO4
Add ddH2O to 1 L - Medium
- 1 L NGM
20 g agar
2.5 g peptone
3 g NaCl
5 mg/ml cholesterol in 1 ml ethanol
Autoclave at 121 °C for 20 min, after autoclave, add:
1 ml 1 M MgSO4 (sterilized)
1 ml 1 M CaCl2 (sterilized)
1 ml 1 M phosphate buffer pH 6.0 (sterilized) - NGM-AG
NGM with 100 µg/ml ampicillin and 50 µg/ml gentamicin - NGMY
NGM plus yeast extract 20 g/L - NGMY liquid
NGMY without agar - NGMS
NGM-AG plus D-sorbitol - 15 mM ATA solution (autoclave)
0.634 g aurintricarboxylic acid in 100 ml ddH2O and autoclave at 121 °C for 20 min - 50 mM NaCl solution (autoclave)
5.8 g NaCl in 2 L ddH2O and autoclave at 121 °C for 20 min - CaCl2 1700 (500 ml, autoclave at 121 °C for 20 min)
15.0 g CaCl2
17.5 g NaCl - Lysis buffer
CaCl2 1700:C4 Caylase = 10 ml:100 mg (or CaCl2 1700:lysing enzyme = 10 ml:200 mg)
Note: Prepare fresh every time, filter with syringe and film. - STC 1700 (500 ml, autoclave)
109.3 g sorbitol
5 ml Tris/HCl
22.8 g CaCl2
1 g NaCl - PEG 1700 (100 ml, autoclave)
60 g PEG 4000
1 ml Tris/HCl
0.74 g CaCl2·2H2O
Note: Tris/HCl solution stored at 1 M pH 7.5, 60.6 g/500 ml, MW = 121.2.
Acknowledgments
We thank Dr. Eric Record for his help in establishing the protocol and Carole Couillault and Zhang Xing for their assistance. We also thank Dr. Nathalie Pujol for her input and suggestions. This work was supported by a program grant from the ANR (ANR-12-BSV3-0001-01) and institutional funding from INSERM, CNRS and AMU. Le He was supported by the Chinese Scholarship Council.
References
- Lebrigand, K., He le, D., Thakur, N., Arguel, M. J., Polanowska, J., Henrissat, B., Record, E., Magdelenat, G., Barbe, V., Raffaele, S., Barbry, P. and Ewbank, J. J. (2016). Comparative genomic analysis of Drechmeria coniospora reveals core and specific genetic requirements for fungal endoparasitism of nematodes. PLoS Genet 12(5): e1006017.
- Ochman, H., Gerber, A. S. and Hartl, D. L. (1988). Genetic applications of an inverse polymerase chain reaction. Genetics 120(3): 621-623.
- Turgeon, B. G., Condon, B., Liu, J. and Zhang, N. (2010). Protoplast transformation of filamentous fungi. In: Sharon, A. (Ed.). Molecular and cell biology methods for fungi. Humana pp: 3-19.
- Stiernagle, T. (2006). Maintenance of C. elegans. In: The C. elegans Research Community (Ed.). WormBook. WormBook.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
He, L. D. and Ewbank, J. J. (2017). Polyethylene Glycol-mediated Transformation of Drechmeria coniospora. Bio-protocol 7(5): e2157. DOI: 10.21769/BioProtoc.2157.
Category
Microbiology > Microbial genetics > Transformation
Molecular Biology > DNA > Transformation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









