- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
An HPLC-based Method to Quantify Coronatine Production by Bacteria
Published: Vol 7, Iss 5, Mar 5, 2017 DOI: 10.21769/BioProtoc.2147 Views: 9266
Reviewed by: Jyotiska ChaudhuriHonghong WuDe Michele Roberto

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An HPLC-based Assay to Study the Activity of Cyclic Diadenosine Monophosphate (C-di-AMP) Synthase DisA from Mycobacterium smegmatis
Avisek Mahapa [...] Dipankar Chatterji
Dec 20, 2024 1789 Views

Protocol for Inoculation of PGPR Staphylococcus sciuri to Seeds and Seedlings of Rice and Tomato Plants for Increased Root and Shoot Growth
Girija Somna [...] Dinakar Challabathula
Mar 20, 2025 2200 Views

In Silico Prediction and In Vitro Validation of Bacterial Interactions in the Plant Rhizosphere Using a Synthetic Bacterial Community
Arijit Mukherjee [...] Sanjay Swarup
Nov 5, 2025 1671 Views
Abstract
Coronatine is a polyketide phytotoxin produced by several pathovars of the plant pathogenic bacterium Pseudomonas syringae. It is one of the most important virulence factors determining the success of bacterial pathogenesis in the plant at both epiphytic and endophytic stages of the disease cycle. This protocol describes an optimized procedure to culture bacterial cells for coronatine production and to quantify the amount of coronatine secreted in the culture medium using an HPLC-based method.
Keywords: CoronatineBackground
Coronatine (COR), a potent bacterial phytotoxin, is a molecular mimic of the plant hormone jasmonoyl-L isoleucine (JA-Ile). As such, COR activates jasmonic acid (JA) signaling, induces JA-responsive genes, and antagonizes the action of the immune signal salicylic acid. COR consists of two components, coronafacic acid (CFA) and coronamic acid (CMA). The genes that encode for CMA and CFA biosynthesis are not constitutively expressed in the bacterium. Instead, these genes are induced on the plant leaf surface, in planta or in vitro when the bacterium is grown in inducing medium (Palmer and Bender, 1993; Panchal et al. 2016). This article describes a method adapted from Panchal et al. (2016) to determine the ability of bacteria to produce coronatine, which can be used as an indication of virulence.
Materials and Reagents
- 1.5 ml microfuge tubes (VWR, catalog number: 20170-650 )
- Centrifuge tubes, polypropylene, sterile, 50 ml (Corning, Falcon®, catalog number: 352070 )
- Micropipettes (Mettler-Toledo, Rainin, catalog number: KitPR-START )
- Petri dishes, 100 x 15 mm (VWR, catalog number: 25384-088 )
- Plastic bags
- Square cuvette, 10 mm path length (Thermo Fisher Scientific, catalog number: 331709 )
- Pasteur pipette (VWR, catalog number: 14673-010 )
- Pseudomonas syringae bacterial culture (can be maintained in 25% glycerol at -80 °C)
- Coronatine (Sigma-Aldrich, catalog number: C8115 )
- Glycerol (VWR, BDH®, catalog number: BDH1172-1LP )
- Sterile distilled ultrapure water
- Trifluoroacetic acid (TFA) (Sigma-Aldrich, catalog number: 302031 )
- Acetonitrile (Sigma-Aldrich, catalog number: 34998 )
- Rifampicin
- 3 N HCl (VWR, BDH®, catalog number: BDH7375-1 )
- Ethyl acetate (VWR, BDH®, catalog number: BDH1123-4LG )
- Sodium hydroxide (NaOH) (Fisher Scientific, catalog number: S318-1 )
- BCA Protein Assay Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 23225 )
- Tryptone (IBI Scientific, catalog number: 41116105 )
- Yeast extract (U.S. Biotech Sources, BTS, catalog number: Y01PD-500 )
- Sodium chloride (NaCl) (Fisher Scientific, catalog number: S271-500 )
- Bacteriological agar (IBI Scientific, catalog number: IB49171 )
- Ammonium chloride (NH4Cl) (VWR, catalog number: 470300-196 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (VWR, catalog number: 97062-134 )
- Potassium phosphate (KH2PO4) (VWR, catalog number: 97062-350 )
- Potassium hydrogen phosphate trihydrate (K2HPO4·3H2O) (VWR, catalog number: ALFA33365.A1 )
- Potassium nitrate (KNO3) (VWR, catalog number: 97065-038 )
- Iron(III) chloride (FeCl3) (VWR, catalog number: 470301-584 )
- Glucose (VWR, catalog number: 101094-092 )
- Low-sodium Luria Bertani medium (see Recipes)
- Liquid HSC medium (see Recipes)
Equipment
- HPLC system: Agilent 1200 HPLC (Agilent Technologies, model: Agilent 1200 HPLC) equipped with quaternary pump, autosampler, thermostated column compartment, and a diode array
- C8 column, 4.6 x 250 mm, 5 µm (ASTEC, Whippany, NJ, USA or equivalent)
- Shaker incubator (Eppendorf, model: New BrunswickTM Innova® 42R )
- Spectrophotometer (Thermo Fisher Scientific, Thermo Scientific, model: Spectronic 20D+ )
- Microcentrifuge (Eppendorf, model: 5418 )
- Water bath (Thermo Fisher Scientific, Thermo ScientificTM, model: Precision GP 2S )
- Vortex (VWR, catalog number: 945300 )
- Autoclave (VWR, catalog number: 97002-402 )
- Laminar flow hood (ESCO, model: AC1-4E8 )
- Centrifuge (Eppendorf, model: 5810 )
- Synergy water purification system (EMD Millipore, catalog number: SYNS0HFWW )
Software
- Chromatography data system software (Agilent Technologies, Palo Alto, CA)
- Microsoft Excel
Procedure
- Calibration curve
- Calibration curves must be created using purified COR to determine the exact concentration of COR in biological samples. Prepare five COR solutions with different concentrations (5-50 µg/ml) in 1.5 ml microfuge tubes; so that 5 µl of the solutions represents a range of COR amounts between 25-250 ng. COR is detected by HPLC at 208 nm using a C8 column (4.6 x 250 mm, 5 µm). Isocratic separations are performed using a 0.05% TFA:acetonitrile (60:40 v/v) mobile phase with a flow rate of 1 ml/min. The column temperature is set at 25 °C. Inject 5 µl aliquots of purified COR into the HPLC C8 column. Pure coronatine must produce a peak at 9.4 min.
- Using the chromatography data system software, integrate the area under the peak for every concentration of COR. Plot a graph of peak area versus ng of coronatine injected as shown in Figure 1. Calculate the linear equation of the trend line obtained using linear regression analysis.
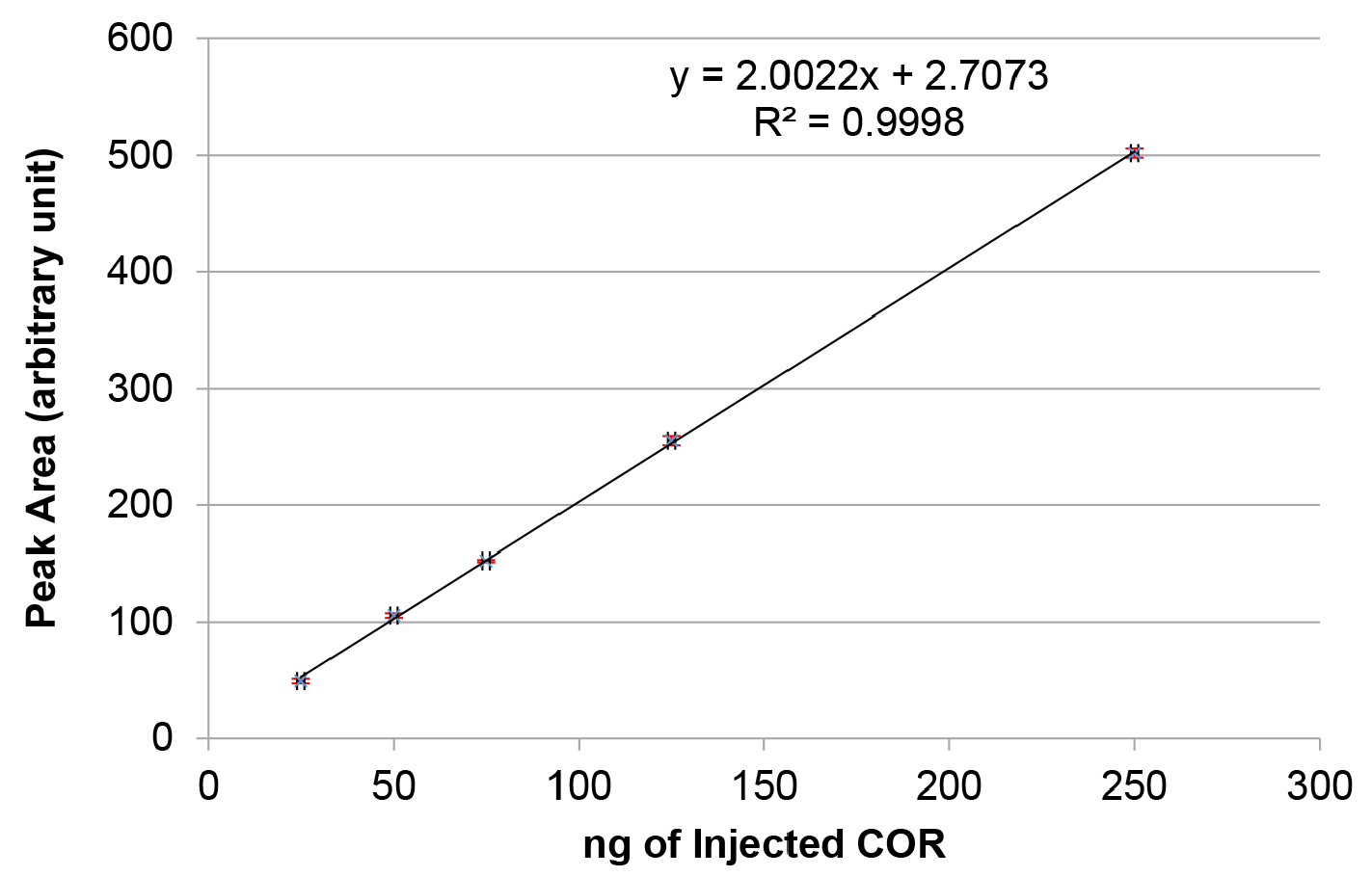
Figure 1. Standard curve for 5-50 μg/ml of coronatine (COR). Purified COR was injected in different concentrations and the peak area (in arbitrary units) for each injection was quantified with the chromatography data system software. Note that the red error bars for each data point are very small and do not clearly appear in the graph.
- Calibration curves must be created using purified COR to determine the exact concentration of COR in biological samples. Prepare five COR solutions with different concentrations (5-50 µg/ml) in 1.5 ml microfuge tubes; so that 5 µl of the solutions represents a range of COR amounts between 25-250 ng. COR is detected by HPLC at 208 nm using a C8 column (4.6 x 250 mm, 5 µm). Isocratic separations are performed using a 0.05% TFA:acetonitrile (60:40 v/v) mobile phase with a flow rate of 1 ml/min. The column temperature is set at 25 °C. Inject 5 µl aliquots of purified COR into the HPLC C8 column. Pure coronatine must produce a peak at 9.4 min.
- Determining bacterial production of COR
- From the glycerol stock, streak the bacterial culture on low-salt Luria Bertani (LS-LB) medium supplemented with appropriate antibiotic and incubate the medium plate at 28 °C for approximately 30 h. For example, 100 µg/ml rifampicin is used to culture COR-producing Pseudomonas syringae pv. tomato DC3000.
- Inoculate a small part of a single colony (just touch with tip) from this fresh plate into 10 ml of LS-LB medium with appropriate antibiotic in a 50 ml tube and incubate at 28 °C and 200 rpm. Grow until mid to late log phase is reached (OD600 of 0.8). It will take approximately 10-12 h. Check optical density (OD) at 600 nm using a spectrophotometer.
- Centrifuge the bacterial suspension at 2,600 x g for 20 min in 50 ml tube. Remove the supernatant and re-suspend the pellets in 10 ml of sterile water. Vortex thoroughly to wash the cells. Vortex continuously at full speed until the cells are completely re-suspended and there is no visible cell clumps. Repeat this step twice. Finally, re-suspend the total bacterial cells in 40 ml sterile water to obtain a 0.2 OD600 inoculum that corresponds to 1 x 108 cells for Pseudomonas syringae pv. tomato. The calculated approximation of the OD600 is sufficient.
- Transfer 1 ml from this culture to 10 ml of HSC medium in a 50 ml tube and incubate for 24 h at 18 °C and 200 rpm. More volume of culture can be used if greater concentrations of COR are required. However, 1 ml is adequate for COR detection. From 10 ml of bacterial culture, one can obtain a concentration of COR in the range of 0.5-2 µg/mg of total protein.
- After 24 h, transfer 0.6 ml of culture to microfuge tubes. Spin at a maximum speed (or 16,000 x g) at room temperature in a microcentrifuge for 3 min to pellet cells.
- Transfer 0.5 ml of supernatant to fresh microfuge tubes and store pelleted cells at -80 °C. These cells are used to estimate total protein content as described in step B14 below.
- To the supernatant, add 25 µl of 3 N HCl and 0.5 ml ethyl acetate to each tube. Vortex the tubes briefly for 30 sec at maximum speed. Allow phases to separate. The phases should separate out within two minutes.
- Transfer 0.4 ml of the ethyl acetate phase (upper layer) to a fresh microfuge tube.
- Repeat steps B7-B8 twice (Figure 2).
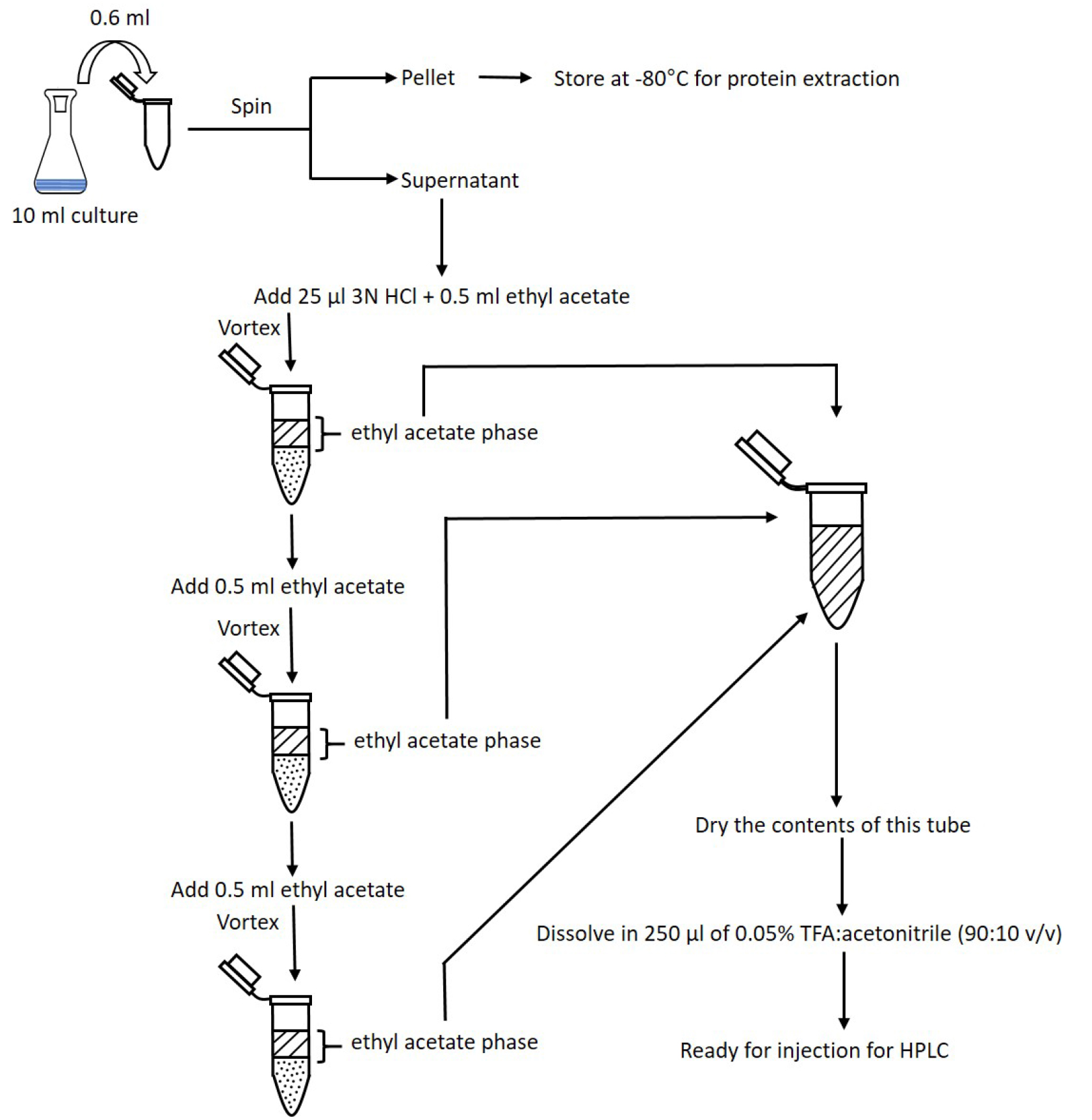
Figure 2. Flowchart of the COR extraction. Steps for extracting COR from bacterial culture supernatant using ethyl acetate. The three ethyl acetate phases are combined in a single tube and let dry. Dried COR is dissolved in TFA:acetonitrile and this solution is now ready for injection into HPLC. - Dry contents of tubes under a stream of compressed air. One method is to float tubes in a 55 °C water bath, using a Pasteur pipette to gently blow air into each tube (see Figure 3 for a representative setup). Alternatively, tubes may be left open on the bench at room temperature overnight.
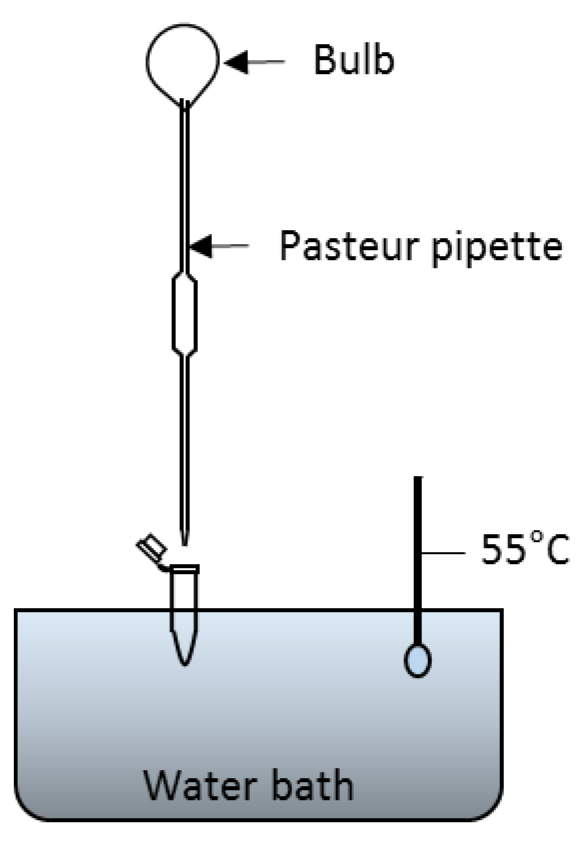
Figure 3. Set up for drying the contents of microfuge tubes in a water bath. After COR is extracted from bacterial culture with ethyl acetate, the ethyl acetate is evaporated by floating the tube on water bath set at 55 °C and blowing air using Pasteur pipette. Alternatively, the tubes can be left open to dry overnight. - For HPLC analysis, add 250 µl of 0.05% TFA:acetonitrile (90:10 v/v) to each dry tube. Vortex each tube for 15 sec. Samples are now ready for direct injection into the HPLC system.
- Determine the presence of COR by HPLC using a C8 column (4.6 x 250 mm, 5 µm) with detection at 208 nm. Isocratic separations are performed using a 0.05% TFA:acetonitrile (60:40 v/v) mobile phase with a flow rate of 1 ml/min. The injection volume is 5 µl and the column temperature is set at 25 °C.
- Inject the unknown sample into the HPLC system. If a peak is observed at 9.4 min, determine the area under the peak. Using the peak area, apply the standard curve and linear equation to determine the ng of coronatine injected in 5 µl. Then, calculate µg/ml coronatine for the sample.
- The amount of COR produced can be expressed as a function of protein concentration. The cells obtained in step B6 are lysed by suspending the pellet in 1 ml of 1 N NaOH followed by boiling and freezing three times. The total protein content in bacterial cell lysates is determined with the Pierce BCA Protein Assay Kit according to manufacturer’s instructions. Express COR concentration as µg/mg protein as shown by Panchal et al., (2016).
- From the glycerol stock, streak the bacterial culture on low-salt Luria Bertani (LS-LB) medium supplemented with appropriate antibiotic and incubate the medium plate at 28 °C for approximately 30 h. For example, 100 µg/ml rifampicin is used to culture COR-producing Pseudomonas syringae pv. tomato DC3000.
Data analysis
- The COR-producing bacterium P. syringae pv. tomato strain DC3000 and its derivative COR-defective mutant strains DC3118 or DB29 must be used as positive and negative controls for this assay, respectively.
- At least three technical replicates and three biological replicates should be used for quantification of COR. Statistical analysis should be done by calculating the average and standard error of COR production across replicates. Significance of the difference between two samples can be obtained by performing a Student’s t-test. These analyses and linear regression can be done with Microsoft Excel or other comparable statistical analysis software.
Notes
- Always use bacterial pathogen isolates that have been freshly streaked from glycerol stock. Multiple sub-culturing of isolates may lead to loss of virulence and COR production. Ideally, the bacterium pathogen is isolated from the plant host every two years.
- At least 24 h of incubation in HSC medium is required for consistent production of COR.
Recipes
- Low-sodium Luria Bertani medium
10 g/L tryptone
5 g/L yeast extract
5 g/L NaCl
2.5% agar
Adjust pH to 7.0 with NaOH
Autoclave medium at 15 psi, 120 °C for 15 min. Allow the medium to cool down to about 55 °C and add appropriate antibiotic as needed. Pour medium into plates in a laminar flow hood. Store plates in plastic bags at 4 °C to avoid medium dehydration - Liquid HSC medium
Part A:
1.0 g NH4Cl
0.2 g MgSO4·7H2O
4.1 g KH2PO4
3.6 g K2HPO4·3H2O
0.3 g KNO3
10 ml 2 mM FeCl3
890 ml ddH2O
Adjust pH to 6.8
Autoclave at 10 psi, 120 °C for 20 min
Part B:
20 g glucose
100 ml ddH2O
Filter-sterilize the solution
Store parts A and B separately at 4 °C. Mix part A and part B (9:1 ratio) right before use
Acknowledgments
We thank Dr. Carol Bender for useful advice on the coronatine extraction method. This study was supported by a grant from the US National Institute of Allergy and Infectious Disease (5R01AI068718) to Dr. Maeli Melotto.
References
- Palmer, D. A. and Bender, C. L. (1993). Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl Environ Microbiol 59(5): 1619-1626.
- Panchal, S., Roy, D., Chitrakar, R., Price, L., Breitbach, Z. S., Armstrong, D. W. and Melotto, M. (2016). Coronatine facilitates Pseudomonas syringae infection of Arabidopsis leaves at night. Front Plant Sci 7: 880.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Panchal, S., Breitbach, Z. S. and Melotto, M. (2017). An HPLC-based Method to Quantify Coronatine Production by Bacteria. Bio-protocol 7(5): e2147. DOI: 10.21769/BioProtoc.2147.
Category
Microbiology > Microbial biochemistry > Other compound
Plant Science > Plant immunity > Host-microbe interactions
Cell Biology > Cell isolation and culture > Cell isolation
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










