- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Expression and Purification of the GRAS Domain of Os-SCL7 from Rice for Structural Studies
Published: Vol 7, Iss 3, Feb 5, 2017 DOI: 10.21769/BioProtoc.2122 Views: 8536
Reviewed by: Arsalan DaudiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Streamlining Protein Fractional Synthesis Rates Using SP3 Beads and Stable Isotope Mass Spectrometry: A Case Study on the Plant Ribosome
Dione Gentry-Torfer [...] Federico Martinez-Seidel
May 5, 2024 2869 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1989 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1772 Views
Abstract
GRAS proteins, named after the first three members GAI, RGA and SRC, has been found in 294 embryophyta species and is represented by 1,035 sequences. They belong to a plant-specific protein family and play essential roles in plant growth and development. Proteins in this family are defined as minimally containing a conserved GRAS domain, which is about 350-450 resides and can be subdivided into five distinct motifs with their name derived from the most prominent amino acids: LRI (leucine-rich region I), VHIID, LRII (leucine-rich region II), PFYRE and SAW and mainly function in the interaction between GRAS proteins and their partners (Sun et al., 2012).By phylogenetic analysis, the GRAS family can be divided into more than ten subfamilies, of which SCL4/7 is one important subgroup and functions in response to environmental stresses. Here we describe a detailed protocol for the expression and purification of the GRAS domain of Os-SCL7, a SCL4/7 member in rice, which enables us to crystallize it and determine its structure.
Keywords: ExpressionBackground
The GRAS proteins are a large family that plays vital roles in plant development and signaling transduction. Findings indicate that some family members such as DELLAs function as a repressor of GA responsive plant growth and are key regulatory targets in the GA signaling pathway (Murase et al., 2008), NSP1 and NSP2 play important roles in regulating nodulation development and signaling (Kaló et al., 2005), the proteins SCR and SHR together play an important role in the control of radial patterning for both the root and shoot (Helariutta et al., 2000), AtLAS is a key regulator in the developmental processes of the axillary meristem (Greb et al., 2003), HAM functions in shoot meristem maintenance (Stuurman et al., 2002).
Based on sequence analyses, GRAS proteins include a variable N-terminal domain and a widely and highly conserved C-terminal domain known as the GRAS domain. The N-terminal domains constitute a plant-specific unfoldome and may act as molecular bait by initiating the key molecular recognition events (Uversky et al., 2010). And the C-terminal GRAS domain is highly conserved in the whole GRAS family, suggesting that these proteins share a similar function and/or a common mode-of-action (Sun et al., 2012).
Though many members of GRAS proteins have been studied, the functional mechanism of GRAS proteins is still unclear. Structural descriptions of GRAS proteins may deeply clarify the functional mechanism of this family. However as yet little structural analyses have been reported, mainly due to the difficulties in obtaining sufficient quality and quantity of GRAS proteins. Soto et al. (2014) reported the expression and purification of the GRAS domain of rice SLR1. They constructed a GST-SLR1 fusion protein and expressed it in E. coli. But the expression levels were low (0.5 mg [TB medium] or 0.2 mg [M9 medium] of purified protein from 1 L flask culture). With some modifications, they obtained 1-3 mg of stable isotope labeled purified protein at 87% purity from 1 L of fermenter culture. However, the expression levels and purity of SLR1 are both not enough for crystallization. Moreover, reducing the protein synthesis rate by low culture temperature and speed, co-expressing with chaperonins, expressing as a fusion protein with soluble tags such as glutathione S-transferase, thioredoxin and maltose-binding protein or mutating some hydrophobic or disulfide forming amino acids are usually used to improve the expression and solubility of target proteins. While using more purification methods and steps will help to improve the purity of target proteins. In order to get a high purity and quantity of GRAS protein, we established a protocol for easy expression and purification of the GRAS domain of Os-SCL7 from rice.
Materials and Reagents
- Amicon Ultra centrifugal filters, 30 K (Merck Millipore, catalog number: UFC903096 )
- Amicon Ultra centrifugal filters 30,000 MWCO (EMD Millipore, catalog number: UFC803024 )
- Economical biotech dialysis membrane, 14 KD (Sangon Biotech, catalog number: TX0111 )
- Escherichia coli (E. coli) (BL21) (Thermo Fisher Scientific, InvitrogenTM, catalog number: C6000-03 )
- pET32aM (Novagen, modified from pET32a by inserting a TEV protease cut site inside the NcoI cleavage site) (Figure 1)
- Rice (NM_001057650, residues 201-578)
- Ampicillin (Sangon Biotech, catalog number: A100741 )
- Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Sangon Biotech, catalog number: A100487 )
- Chelating Sepharose Fast FLow (GE Healthcare, catalog number: 17057502 )
- Nickel(II) sulfate hexahydrate (NiSO4·6H2O) (Sangon Biotech, catalog number: A600658 )
- Imidazole (Sangon Biotech, catalog number: A500529 )
- TEV (tobacco etch virus) protease (produced in-house) (Fang et al., 2007)
- Tryptone (Oxoid, catalog number: LP0042 )
- Yeast extract (Oxoid, catalog number: LP0021 )
- Sodium chloride (NaCl) (Sangon Biotech, catalog number: A501218 )
- Agar (Oxoid, catalog number: LP0011 )
- Tris-base (Sangon Biotech, catalog number: A100826 )
- Hydrochloric acid (HCl) (Sinopharm Chemical Reagent, catalog number: 7647-01-0 )
- Glycerol (Sangon Biotech, catalog number: A100854 )
- Ethylenediaminetetraacetic acid disodium salt (EDTA) (Sangon Biotech, catalog number: A100105 )
- β-mercaptoethanol (AMRESCO, catalog number: 60-24-2 )
- Tween-20 (Sangon Biotech, catalog number: A100777 )
- Ethanol anhydrous (Sangon Biotech, catalog number: A500737 )
- Phosphoric acid ortho (85%) (Sangon Biotech, catalog number: A502803 )
- Acetic acid (Sangon Biotech, catalog number: A501931 )
- Coomassie Brilliant Blue G-250 (Sangon Biotech, catalog number: A100615 )
- Coomassie Brilliant Blue R-250 (Sangon Biotech, catalog number: A100472 )
- SDS (Sangon Biotech, catalog number: A100227 )
- Isopropanol (Sangon Biotech, catalog number: A507048 )
- LB medium (see Recipes)
- LB-agar plates (see Recipes)
- Lysis buffer (see Recipes)
- Chromatography buffer (see Recipes)
- Coomassie Brilliant Blue G-250 buffer (see Recipes)
- SDS-PAGE staining solution (see Recipes)
- SDS-PAGE destaining solution (see Recipes)
Figure 1. The map of pET32aM plasmid
Equipment
- 2 L flask
- Spectrophotometer device (Kebo instrument, model: UV-1100 )
- Sonicator (Scientz, model: IID )
- Refrigerated centrifuge (Eppendorf, model: 5810 R )
- Chromatography system (GE Healthcare, model: INV-907 )
Note: This product has been discontinued. - HiLoad 16/600 Superdex 200(GE Healthcare, catalog number: 28-9893-35 )
- Electrophoresis system (Liuyi, model: DYY-6D )
Procedure
- Clone the gene encoding the GRAS domain Os-SCL7 from rice (NM_001057650, residues 201-578) into the modified pET32a vector (pET32aM) (Figure 1).
- Transform the recombinant plasmid (OsSCL7201-578-pET32aM), which was verified by DNA sequencing, into Escherichia coli strain BL21 (DE3) by heat shock method (Hanahan, 1983) and plate on LB-agar plates containing 100 mg/L ampicillin for 12 h at 37 °C.
- Pick a single colony and culture the E. coli in 10 ml LB medium containing 100 mg/L ampicillin at 37 °C for 5 h. Then transfer 10 ml culture to 1 L fresh LB medium containing 100 mg/L ampicillin (in a 2 L flask) and culture at 37 °C until OD600 nm reaches 0.4-0.6.
- Cool the cultures to about 16 °C and then induce protein expression with 0.3 mM IPTG for 12-14 h at 16 °C, 180 rpm.
- Harvest the cells by centrifugation at 8,800 x g for 5 min and discard the supernatant, then suspend the cells in 50 ml lysis buffer by vortexing.
- Sonicate the resuspended cells on ice with the power of 380 W for 30 min (ultrasonication is set to 2 sec ‘on’ and 6 sec ‘off’, and repeat this cycle).
Note: The proteins should always be kept on ice and avoided of high energy produced during the ultrasonicaton process. Having an interval time should be better in this process (here the ultrasonication process is set as three times with 10 min for each time and 2 min interval between every two times). - After sonication, centrifuge the sample at 18,000 x g for 30 min.
- Transfer the supernatant to a new tube, add imidazole into the supernatant to a final concentration of 20 mM and mix it thoroughly. Keep the pellet (containing the cell debris and protein aggregates) on ice before being subjected to SDS-PAGE (step 11).
Note: Adding a final concentration of 20 mM imidazole into the supernatant can effectively reduce non-specific proteins binding. - Load the sample onto a nickel-sepharose affinity resin (Chelating sepharose Fast FLow) which was pre-equilibrated with lysis buffer.
- Elute the bound protein with lysis buffers containing a gradient of imidazole (10 mM, 20 mM, 50 mM and 500 mM).
- Subject the eluted protein mixed with loading buffer to SDS-PAGE (Figure 2a). The protein of interest should be between 45 kDa and 66.2 kDa, according to the molecular weight 59 kDa of OsSCL7 GRAS domain fused with TRX and 6His-tag.
Note: In order to get high purity proteins, it’s better to use a series concentration of imidazole to wash the nonspecific binding proteins and make sure each concentration of imidazole could not nearly elute any protein (see Notes). The target proteins are in the 500 mM imidazole eluent. - Add TEV protease to the eluted proteins of interest (mTEV:mprotein = 1:10) to remove the TRX and 6His-tag from OsSCL7 GRAS domain. Then transfer the mixture to a dialysis membrane and dialyze it in lysis buffer on a rotator overnight at 4 °C.
Note: In this step, the lysis buffer is used to dialyze imidazole to a low concentration. Generally it is lower than 5 mM. - Load the cleaved protein onto the nickel-sepharose affinity resin again. The target proteins are in the flow through and subsequent 20 mM imidazole eluent. The band of the target protein is between 35 and 45 kDa, according to the molecular weight 42 kDa of the OsSCL7 GRAS domain (Figure 2b).
- Then load the sample into a HiLoad 16/600 Superdex 200 column pre-equilibrated with 1.2 column volume of chromatography buffer (Figure 3a). The samples of OsSCL7 monomer and dimer were assessed by SDS-PAGE (Figure 3b).
- Pool the fractions containing target protein and concentrate the sample to 5 to 8 mg/ml, measured by Bradford method on spectrophotometer device (Bradford, 1976), with Amicon Ultra centrifugal filters for subsequent crystallization experiment.
Data analysis
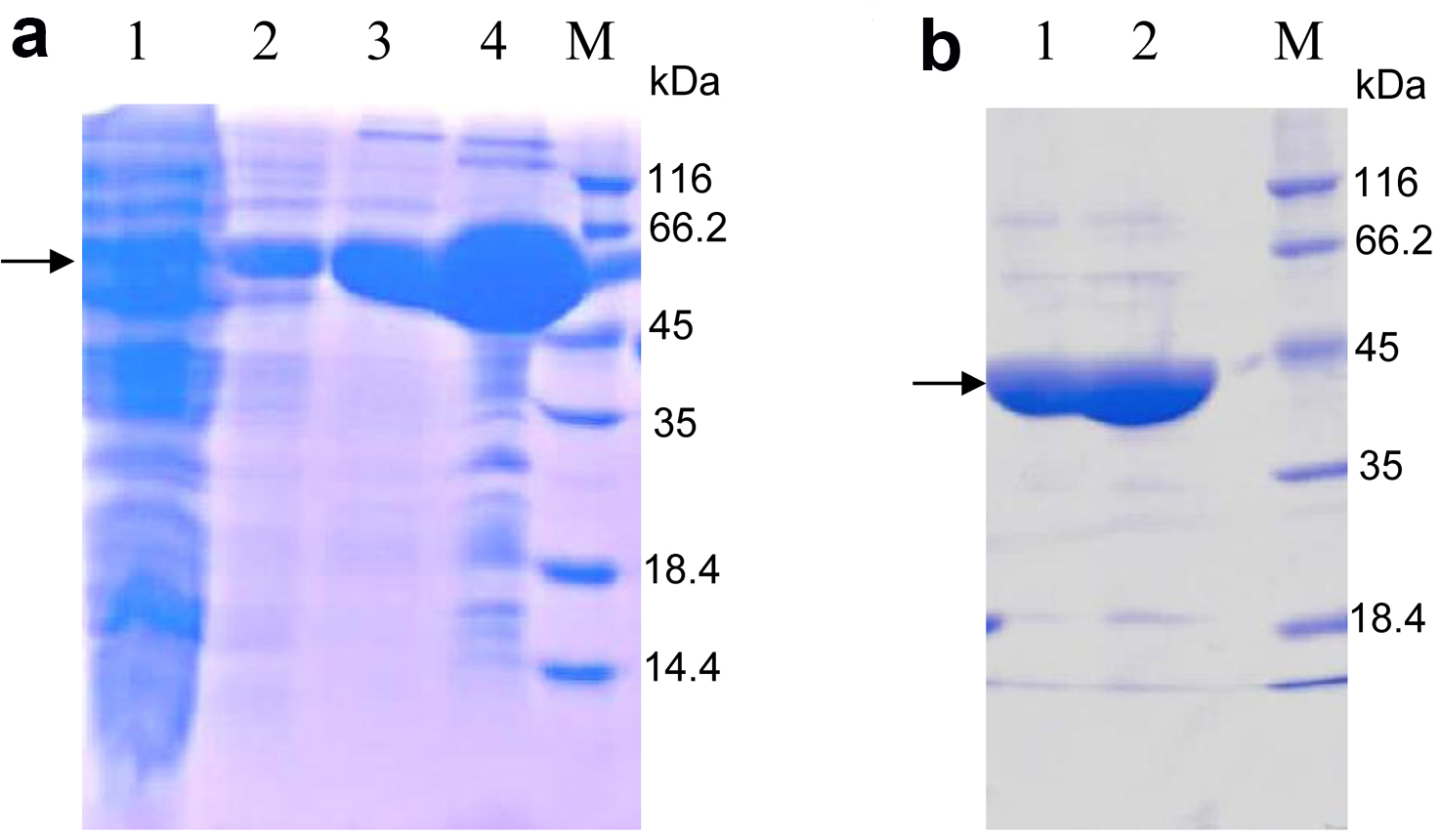
Figure 2. 15% SDS-PAGE analysis of purified OsSCL7 GRAS domain by nickel-sepharose affinity resin stained with Coomassie Brilliant Blue. a. Lane 1: The aggregates (produced in step 8); Lane 2: The flow (step 8); Lane 3: Eluted with 20 mM imidazole; Lane 4: Eluted with 500 mM imidazole; M: Marker. The arrow indicates the similar MW with the predicted MW (59 kDa) of OsCL7 GRAS domain (41.5 kDa) + Trx-tag (17.5 kDa); b. Lane 1: The flow of the second nickel-sepharose affinity resin purification (produced in step 11, The TRX and 6His-tag were cut off from the OsSCL7 GRAS domain), according to the MW of OsSCL7 GRAS domain, around 42 kDa; Lane 2: Elution with 20 mM imidazole, which also contains the OsSCL7 GRAS domain; M: Marker.
Figure 3. Purification of OsSCL7 GRAS domain by size-exclusion chromatography. a. Size-exclusion chromatography (HiLoad 16/600 Superdex 200 column), the dimer and monomer of OsSCL7 GRAS domain appear at 72.26 and 79.68 ml respectively, their corresponding molecular weights are 84 kDa and 42 kDa. b. 15% SDS-PAGE analysis of purified OsSCL7 GRAS domain from fractions of the observed peak in size-exclusion chromatography, showing high purity.
Notes
- The TEV protease prepared here must be pure enough (more than 90%) and the second nickel-sepharose affinity resin step is important for OsSCL7-GRAS domain purification.
- A pre-experiment was performed to determine the concentration of imidazole would elute the target protein well.
- A series of different concentrations of imidazole is used for washing the nonspecific binding proteins (the highest concentration of imidazole used in this step must be lower than which is for elution of the target protein). And Coomassie Brilliant Blue G-250 buffer (see Recipes) is used to detect no proteins can be washed down any longer by each concentration of imidazole.
Recipes
- LB medium
10 g/L tryptone
5 g/L yeast extract
10 g/L NaCl - LB-agar plates
10 g/L tryptone
5 g/L yeast extract
10 g/L NaCl
1.5% agar - Lysis buffer
500 mM NaCl
50 mM Tris-HCl buffer (pH 8.0)
5% glycerol
0.2 mM EDTA
2 mM β-mercaptoethanol
0.1% Tween-20 - Chromatography buffer
100 mM NaCl
25 mM Tris-HCl buffer (pH 8.0)
5% glycerol - Coomassie Brilliant Blue G-250 buffer
47.5 ml ethanol anhydrous
100 ml phosphoric acid ortho (85%)
0.1 g G-250
Add ddH2O to 1 L - SDS-PAGE staining solution
300 ml ethanol anhydrous
100 ml acetic acid
1 g Brilliant Blue R-250
Add ddH2O to 1 L - SDS-PAGE destaining solution
300 ml ethanol anhydrous
100 ml acetic acid
Add ddH2O to 1 L
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31500218).
References
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Fang, L., Jia, K. Z., Tang, Y. L., Ma, D. Y., Yu, M. and Hua, Z. C. (2007). An improved strategy for high-level production of TEV protease in Escherichia coli and its purification and characterization. Protein Expr Purif 51(1): 102-109.
- Greb, T., Clarenz, O., Schafer, E., Muller, D., Herrero, R., Schmitz, G. and Theres, K. (2003). Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev 17(9): 1175-1187.
- Hanahan, D. (1983). Studies on transformation of Escherichia coli with plasmids. J Mol Biol 166(4): 557-580.
- Helariutta, Y., Fukaki, H., Wysocka-Diller, J., Nakajima, K., Jung, J., Sena, G., Hauser, M.T. and Benfey, P. N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101(5): 555-567.
- Kaló, P., Gleason, C., Edwards, A., Marsh, J., Mitra, R. M., Hirsch, S., Jakab, J., Sims, S., Long, S. R., Rogers, J., Kiss, G. B., Downie, J. A. and Oldroyd, G. E. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308(5729): 1786-1789.
- Murase, K., Hirano, Y., Sun, T. P. and Hakoshima, T. (2008). Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456(7221): 459-463.
- Sato, T., Miyanoiri, Y., Takeda, M., Naoe, Y., Mitani, R., Hirano, K., Takehara, S., Kainosho, M., Matsuoka, M., Ueguchi-Tanaka, M. and Kato, H. (2014). Expression and purification of a GRAS domain of SLR1, the rice DELLA protein. Protein Expr Purif 95: 248-258.
- Stuurman, J., Jaggi, F. and Kuhlemeier, C. (2002). Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev 16(17): 2213-2218.
- Sun, X., Jones, W. T. and Rikkerink, E. H. (2012). GRAS proteins: the versatile roles of intrinsically disordered proteins in plant signalling. Biochem J 442(1): 1-12.
- Uversky, V. N. (2010). The mysterious unfoldome: structureless, underappreciated, yet vital part of any given proteome. J Biomed Biotechnol 2010: 568068.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Li, S., Zhao, Y. and Wu, Y. (2017). Expression and Purification of the GRAS Domain of Os-SCL7 from Rice for Structural Studies. Bio-protocol 7(3): e2122. DOI: 10.21769/BioProtoc.2122.
Category
Plant Science > Plant biochemistry > Protein > Isolation and purification
Biochemistry > Protein > Expression
Biochemistry > Protein > Structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










