- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Assays for the Detection of Calreticulin Exposure, ATP and HMGB1 Release upon Cell Death
Published: Vol 6, Iss 24, Dec 20, 2016 DOI: 10.21769/BioProtoc.2076 Views: 20006
Reviewed by: Lee-Hwa TaiJustine MarsolierMichael Enos

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of Autophagosomes in Human Fibroblasts Using Cyto-ID® Staining and Cytation Imaging
Barbara Hochecker [...] Jörg Bergemann
Jul 5, 2024 1660 Views

Microfluidic Cultures of Basal Forebrain Cholinergic Neurons for Assessing Retrograde Cell Death by Live Imaging
Srestha Dasgupta [...] Wilma J. Friedman
Jan 5, 2025 1860 Views

Real-time IncuCyte® Assay for the Dynamic Assessment of Live and Dead Cells in 2D Cultures
Arlene K. Gidda [...] Sharon M. Gorski
Feb 5, 2025 2868 Views
Abstract
Accumulating evidence is revealing the essential role of immune system in cancer treatment. Certain chemotherapeutic drugs can potently induce the release of ‘cell death associated molecular patterns’ (CDAMPs), which accompanies cancer cell demise. CDAMPs can engage corresponding receptors on immune cells and stimulate immune responses to achieve long-term tumor control (Ma et al., 2013; Ma et al., 2014; Yang et al., 2015). Among reported CDAMPs, calreticulin (CALR), ATP and HMGB1 are well known for their immune-stimulatory effect. Here we describe the assays that we applied to measure cell death and these CDAMPs. Briefly, cell death can be analyzed by co-staining of 4’,6-diamidino-2-phenylindole (DAPI) with 3,3’-Dihexyloxacarbocyanine Iodide [DiOC6(3)] or Annexin V. CALR exposure on the cell membrane can be detected by flow cytometry. ATP and HMGB1 release can be quantified by luminescence assay and ELISA assay respectively.
Keywords: Cell deathBackground
Lactate dehydrogenase assay and trypan blue staining are traditional methods to examine cell death. We describe here two feasible and economic solutions to detect apoptotic and necrotic cell death by flow cytometry (FCM). DAPI labels cells with disrupted integrity (necrosis), while Annexin V binds to phosphatidylserine (which is externalized upon apoptosis). DiOC6(3) uptake indicates mitochondrial transmembrane potential (MTP) and the collapse of MTP reveals apoptosis. DAPI doesn’t require compensation with phycoerythrin (PE, which is conjugated with Annexin V protein) or DiOC6(3), and therefore show advantage over propidium iodide (PI) in these assays.
CALR is an endoplasmic reticulum protein, and activation of caspase cascades triggers CALR translocation to cytoplasmic membrane. CALR exposure can be detected by immunofluorescent staining with corresponding antibodies, followed by FCM- or microscopy-based detection. Extracellular ATP can be measured by enzymatic activity of Luciferase, while HMGB1 concentration can be detected by sensitive ELISA kits.
Materials and Reagents
- 24 well plates
- 2 ml Eppendorf tubes
- MCA205 fibrosarcoma cells (H2b) were induced by 3-methylcholanthrene in C57BL/6 mice (Shu et al., 1985). The assays described here are applicable for other cell lines
- DMEM high glucose (Thermo Fisher Scientific, GibcoTM, catalog number: 11965092 , or GE Healthcare, HyCloneTM , catalog number: SH30022 )
- Fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 16000044 )
- 100 mM sodium pyruvate solution (Thermo Fisher Scientific, GibcoTM, catalog number: 11360070 )
- HEPES, 1 M buffer solution (Thermo Fisher Scientific, GibcoTM, catalog number: 15630080 )
- Penicillin and streptomycin
- Mitoxantrone (Sigma-Aldrich, catalog number: M6545 )
- Trypsin (0.25%) (Thermo Fisher Scientific, GibcoTM, catalog number: 15050065 )
- 1x PBS, pH 7.4 (Thermo Fisher Scientific, GibcoTM, catalog number: 10010031 )
- DiOC6(3) (3,3’-dihexyloxacarbocyanine Iodide) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: D273 )
- DAPI (4’,6-diamidino-2-phenylindole, dihydrochloride) (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: D1306 )
- CALR antibody (Ab): rabbit monoclonal Ab, clone number: EPR3924 (Abgent, catalog number: AJ1124a ; or Abcam, catalog number: ab92516 ); or rabbit polyclonal Ab (Abcam, catalog number: ab2907 )
- Alexa488 conjugated goat anti-rabbit IgG (H+L) secondary antibody (Thermo Fisher Scientific, Invitrogen, catalog number: A-11008 )
- ATP Bioluminescent Assay Kit (Sigma-Aldrich, catalog number: FL-AA ) or ENLITEN® ATP Assay System (Promega, catalog number: FF2000 )
- Annexin V Apoptosis Detection Kit (BD, catalog number: 559763 )
- HMGB1 ELISA Kit (Tecan Trading, catalog number: ST51011 )
- 1x Annexin V binding buffer (see Recipes)
- ATP assay mix (see Recipes)
- rLuciferase/Luciferin buffer(see Recipes)
Equipment
- Centrifuge
- Water-Jacketed CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: HERAcell® 150i )
- Multi-channel pipette
- SpectraMax L microplate reader (Molecular Device, model: SpectraMax L )
- SpectraMax i3 multi-mode detection platform (Molecular Device, model: SpectraMax i3 )
- AttuneTM NxT flow cytometer (Thermo Fisher Scientific, model: AttuneTM NxT Flow Cytometer )
Note: Alternatively, VICTORTM X multilabel reader (PerkinElmer) and MACSQuant® Analyzer 10 (Miltenyi Biotec) are also suitable.
Software
- Excel (Microsoft Office)
- GraphPad Prism (San Diego, CA, USA)
- FlowJo software (Treestar Inc., Ashland, OR, USA)
Procedure
MCA205 tumor cells are cultured in DMEM supplemented with 10% FBS, 1 mM sodium pyruvate, 10 mM HEPES and 100 U/ml penicillin and streptomycin. Cells are seeded in 24 well plates (5 x 104 cells in 500 µl culture medium per well). The next day, the culture medium is replaced carefully with 500 µl pre-warmed fresh medium (with or without chemotherapeutic drugs). Culture media can be substituted according to the requirements of different cell lines. The recommended dose for mitoxantrone is 1-10 µM.
- Detection of cell death
- At desired time points, the supernatant is collected thoroughly by pipette. Adherent cells are detached and dispersed with trypsin. All cells are then collected to combine with the original supernatant.
- Cells are collected in 2 ml Eppendorf tubes by spinning at 300 x g, 10 min, 4 °C and the supernatant is discarded. Do not disturb the cell pellets while removing the supernatant.
- Cell pellets are resuspended in PBS containing 1 µM DiOC6(3), incubated at 37 °C for 15 min. The stained cells are washed twice with PBS (10 times the initial volume) and then resuspended in 400 µl PBS containing 1 µg/ml DAPI. The samples are ready for cytofluorometry analysis within 30 min (Figure 1).
- Alternatively, cell pellets are washed with PBS twice and resuspended in 100 µl 1x Annexin V binding buffer containing PE-Annexin V (1:50 dilution) and DAPI (1 µg/ml). The cell concentration should not exceed 1 x 106 cells/ml. After incubation for 15 min at room temperature in the dark, 400 µl 1x Annexin V binding buffer are added to each tube. The samples are ready for cytofluorometry analysis within 30 min (Figure 2).
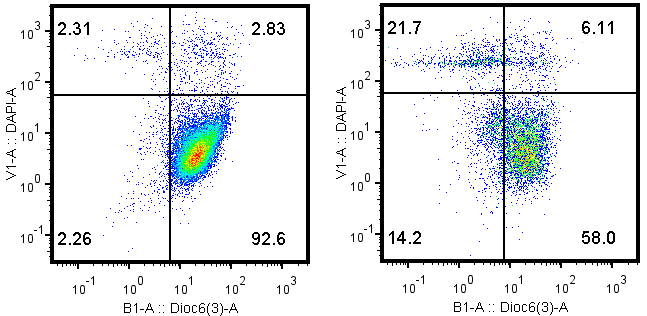
Figure 1. Typical dot plots of DiOC6(3)-DAPI double staining with MCA205 tumor cells that have been pre-treated with PBS control (left) or 1 µM mitoxantrone, 28 h (right)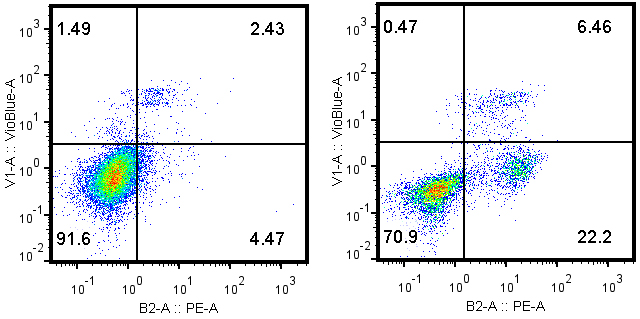
Figure 2. Typical dot plots of Annexin V PE-DAPI double staining with MCA205 tumor cells that have been pre-treated with PBS control (left) or 1 µM mitoxantrone, 24 h (right)
- At desired time points, the supernatant is collected thoroughly by pipette. Adherent cells are detached and dispersed with trypsin. All cells are then collected to combine with the original supernatant.
- Detection of calreticulin (CALR) exposure
- For each sample, the entire well of supernatant and cells are collected and combined in the same Eppendorf tube at desired time points (e.g., 6 h, 16 h, 24 h and 48 h). Discard the supernatant after spinning at 300 x g, 10 min at 4 °C.
- Tumor cells are stained with anti-CALR rabbit monoclonal Ab (1:500 dilution, Abgent, clone EPR3924) or rabbit polyclonal CALR Ab (used at dilution 1:100, Abcam) on ice for 30 min in the dark.
- After a wash with PBS (10 times the initial volume), tumor cells are stained with Alexa488 conjugated goat anti-rat IgG (H+L) secondary antibody (1:500 dilution) on ice for 30 min.
- After wash with PBS (10 times the initial volume), tumor cells are further stained with DAPI at 1 µg/ml, 5 min at room temperature. All samples are kept on ice and analyzed by cytofluorometry within 30 min. To calculate CALR+cells among live cells, DAPI+cells are excluded from the gating (Figure 3).
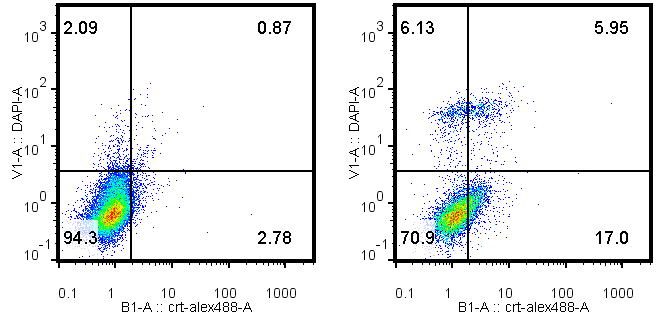
Figure 3. Typical dot plots of CALR-DAPI double staining with MCA205 tumor cells that have been pre-treated with PBS control (left) or 1 µM mitoxantrone, 24 h (right)
- For each sample, the entire well of supernatant and cells are collected and combined in the same Eppendorf tube at desired time points (e.g., 6 h, 16 h, 24 h and 48 h). Discard the supernatant after spinning at 300 x g, 10 min at 4 °C.
- Detection of ATP release
- Three hundred microliters (300 µl) of the supernatant is collected at desired time points and spun at 500 x g, 5 min at 4 °C to remove the inclusion of cells or debris.
- The collected supernatant should be subjected to ATP bioluminescent assay immediately or stored at -80 °C (150 µl per aliquot). Storage of supernatant at room temperature or repeated freeze/thaw leads to rapid ATP degradation. There’s no need to spin supernatant samples again after thawing.
- For ATP Bioluminescent Assay Kit (Sigma-Aldrich), a 25-fold dilution of ATP Assay Mix stock solution with ATP Assay Mix Dilution Buffer is recommended. The samples or standards (100 µl per well) are added into the plate containing diluted assay mix (100 µl per well) with multi-channel pipette rapidly.
- For ENLITEN® ATP Assay System (Promega), the samples and standards (20 µl/well) are added into the plate on ice. Reconstituted rLuciferase/Luciferin Reagent (100 µl/well) are then added into each well with multi-channel pipette rapidly.
- ATP-driven chemiluminescence signals are recorded with a luminescence microplate reader. ATP concentrations in samples are calculated with standard curves (ranging from 10-6 to 10-12 with a 10 fold serial dilution) (Figure 4).
Caution: Microplates should be scanned immediately after samples are mixed with assay mix.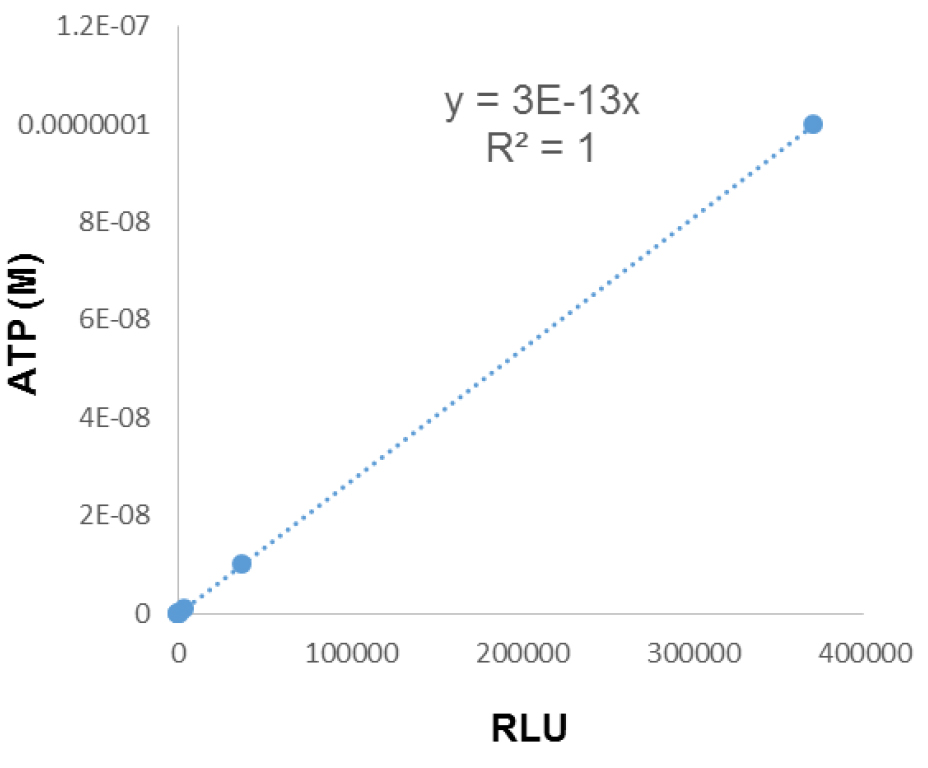
Figure 4. A typical standard curve for calculating ATP content
- Three hundred microliters (300 µl) of the supernatant is collected at desired time points and spun at 500 x g, 5 min at 4 °C to remove the inclusion of cells or debris.
- Detection of HMGB1 release
- Three hundred microliters (300 µl) of the supernatant is collected at desired time points and spun at 500 x g, 5 min at 4 °C to remove the inclusion of cells or debris.
- The supernatant is either applied to ELISA-based quantification freshly, or stored at -80 °C (150 µl per aliquot). Repeated freeze/thaw of samples should be avoided. The concentration of HMGB1 is detected following user’s instruction with standard test procedures.
- Incubation of samples and standards with pre-coated plate at 37 °C for 20-24 h is necessary to reach the maximal sensitivity. High sensitive detection procedures can be applied to detect low HMGB1 content in the samples.
- Optical density is measured with a photometer at 450 nm and 600 nm (used as the reference, subtracted from the 450 nm reading). The concentration of HMGB1 is calculated according to the standard curve (Figure 5).
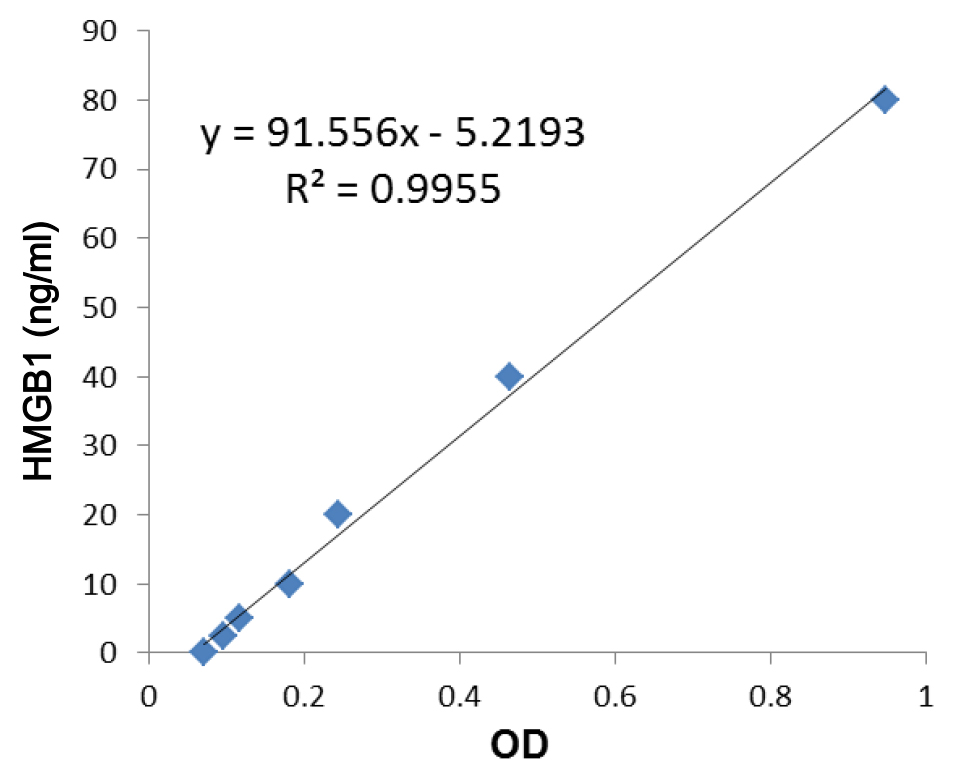
Figure 5. A typical standard curve for calculating HMGB1 content
- Three hundred microliters (300 µl) of the supernatant is collected at desired time points and spun at 500 x g, 5 min at 4 °C to remove the inclusion of cells or debris.
Data analysis
Flow cytometry data can be analyzed by flowJo software (Treestar Inc., Ashland, OR, USA). Apoptotic cells are DAPI-DiOC6(3)low or DAPI-Annexin V+. Necrotic cells are DAPI+DiOC6(3)low or DAPI+Annexin V+. Data should be calculated and presented as mean ± SD (or SEM). Cells with CALR exposure are CALR+DAPI-. The concentration of ATP and HMGB1 are calculated according to standard curves. It is recommended that all experiments should be performed at least 2-3 times independently. Statistics and data visualization are generated by Graphpad Prism software (San Diego, CA, USA). Unpaired, two-tailed Student’s t-test is the preferred method.
Notes
- To compare the cytotoxicity of drugs or the susceptibility of different cells, cell death can be checked at various time points, or in response to different doses of drugs.
- To accurately detect ATP in the supernatant, all procedures should be performed on ice as quickly as possible.
- It is possible to detect ATP from frozen supernatant, which should have been stored at -80 °C preferentially. However, special attentions should be paid during supernatant collection. Cell contamination in the supernatant will give extremely high level of ATP, due to cell rupture after freeze/thaw. The tips of pipette should not reach the bottom of culture plate. Prevent disturbances of the media and collect the supernatant gently.
- Annexin V-PE is more sensitive and reliable than Annexin V-FITC kits to detect cell death.
Recipes
- 1x Annexin V binding buffer
Dilute 10x Annexin V binding buffer provided in Annexin V Apoptosis Detection Kit with ddH2O - ATP assay mix
For ATP Bioluminescent Assay Kit, ATP assay mix and ATP assay mix dilution buffer are dissolved with 5 ml and 50 ml ddH2O respectively on ice for 30 min. Dilute the ATP assay mix stock solution with 25-fold dilution buffer. - rLuciferase/Luciferin buffer
For ENLITEN® ATP Assay System, rLuciferase/Luciferin is reconstituted with 12 ml reconstitution buffer provided in the kit.
Acknowledgments
Dr. Yuting Ma is supported by Chinese National Thousand Young Talents Program, Chinese Academy of Medical Sciences research grant (Grant No. 2015RC310003) and Natural Science Foundation of Jiangsu Province, China (Grant No. BK20160379).
References
- Ma, Y., Adjemian, S., Mattarollo, S. R., Yamazaki, T., Aymeric, L., Yang, H., Portela Catani, J. P., Hannani, D., Duret, H., Steegh, K., Martins, I., Schlemmer, F., Michaud, M., Kepp, O., Sukkurwala, A. Q., Menger, L., Vacchelli, E., Droin, N., Galluzzi, L., Krzysiek, R., Gordon, S., Taylor, P. R., Van Endert, P., Solary, E., Smyth, M. J., Zitvogel, L. and Kroemer, G. (2013). Anticancer chemotherapy-induced intratumoral recruitment and differentiation of antigen-presenting cells. Immunity 38(4): 729-741.
- Ma, Y., Mattarollo, S. R., Adjemian, S., Yang, H., Aymeric, L., Hannani, D., Portela Catani, J. P., Duret, H., Teng, M. W., Kepp, O., Wang, Y., Sistigu, A., Schultze, J. L., Stoll, G., Galluzzi, L., Zitvogel, L., Smyth, M. J. and Kroemer, G. (2014). CCL2/CCR2-dependent recruitment of functional antigen-presenting cells into tumors upon chemotherapy. Cancer Res 74(2): 436-445.
- Shu, S. Y. and Rosenberg, S. A. (1985). Adoptive immunotherapy of newly induced murine sarcomas. Cancer Res 45(4): 1657-1662.
- Yang, H., Yamazaki, T., Pietrocola, F., Zhou, H., Zitvogel, L., Ma, Y. and Kroemer, G. (2015). STAT3 inhibition enhances the therapeutic efficacy of immunogenic chemotherapy by stimulating type 1 interferon production by cancer cells. Cancer Res 75(18): 3812-3822.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ma, Y. and Yang, H. (2016). In vitro Assays for the Detection of Calreticulin Exposure, ATP and HMGB1 Release upon Cell Death. Bio-protocol 6(24): e2076. DOI: 10.21769/BioProtoc.2076.
Category
Cancer Biology > Cell death > Immunological assays
Biochemistry > Other compound > Nucleoside triphosphate
Cell Biology > Cell viability > Cell death
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link