- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of ATP Hydrolytic Activity of Plasma Membrane H+-ATPase from Arabidopsis thaliana Leaves
Published: Vol 6, Iss 23, Dec 5, 2016 DOI: 10.21769/BioProtoc.2044 Views: 9966
Reviewed by: Dennis NürnbergAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1474 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3209 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1824 Views
Abstract
Plant plasma membrane H+-ATPase, which is a P-type ATPase, couples ATP hydrolysis to H+ extrusion and thereby generates an electrochemical gradient across the plasma membrane. The proton gradient is necessary for secondary transport, cell elongation, and membrane potential maintenance. Here we describe a protocol for measurement of the ATP hydrolytic activity of the plasma membrane H+-ATPase from Arabidopsis thaliana leaves.
Keywords: Arabidopsis thalianaBackground
Determination of the plasma membrane H+-ATPase activity is important to elucidate its function and regulatory mechanism. However, it is sometimes difficult to determine the ATP hydrolytic activity of the plasma membrane H+-ATPase, because plant cells contain many ATP hydrolytic enzymes. This protocol is developed based on the publications by Uemura and Yoshida (1986) and Kinoshita et al. (1995). We used KNO3 as an inhibitor of V-type ATPases, ammonium molybdate as an inhibitor of acid phosphatases, oligomycin as an inhibitor of F-type ATPases, and NaF as an inhibitor of phosphatases (Shimazaki and Kondo,1987; Kinoshita et al.,1995). Orthovanadate inhibits the P-type ATPase and thus can be used to measure the activity of the plasma membrane H+-ATPase by assessing the vanadate-sensitive Pi release from ATP hydrolysis. The released Pi reacts with molybdate to form a blue complex which can then be quantified by measuring absorption at 750 nm.
Materials and Reagents
- Ultracentrifuge tube (Beckman Coulter, catalog number: 349623 )
- Cuvette (100 µl) (Beckman Coulter, catalog number: 523270 )
Note: This product has been discontinued. - Arabidopsis thaliana ecotype Col-0
- Dithiothreitol (DTT) (NACALAI TESQUE, catalog number: 14128-04 )
- Phenylmethylsulfonyl fluoride (PMSF) (NACALAI TESQUE, catalog number: 273-27 )
- Leupeptin (Wako Pure Chemical Industries, catalog number: 126-03754 )
- MOPS (NACALAI TESQUE, catalog number: 23415-54 )
- Oligomycin (Sigma-Aldrich, catalog number: 75351 )
- Sodium chloride (NaCl) (Wako Pure Chemical Industries, catalog number: 191-01665 )
- Ethylenediamine-N,N,N’,N’-tetraacetic acid (EDTA) (Dojindo Molecular Technologies, catalog number: N001-10 )
- Sodium fluoride (NaF) (NACALAI TESQUE, catalog number: 31420-82 )
- Tris (NACALAI TESQUE, catalog number: 35406-91 )
- 2-(N-morpholino)ethanesulfonic acid (MES) (NACALAI TESQUE, catalog number: 21623-26 )
- Magnesium sulfate (MgSO4) (Wako Pure Chemical Industries, catalog number: 131-00405 )
- Potassium chloride (KCl) (Wako Pure Chemical Industries, catalog number: 163-03545 )
- Potassium nitrate (KNO3) (Wako Pure Chemical Industries, catalog number: 160-04035 )
- Ammonium molybdate (Wako Pure Chemical Industries, catalog number: 016-06902 )
- Triton X-100 (Wako Pure Chemical Industries, catalog number: 169-21105 )
- ATP (NACALAI TESQUE, catalog number: 10406-61 )
- Sodium orthovanadate (Sigma-Aldrich, catalog number: S6508 )
- SDS (NACALAI TESQUE, catalog number: 31607-65 )
- Sodium molybdate (Wako Pure Chemical Industries, catalog number: 196-02472 )
- Sulfuric acid (H2SO4) (Wako Pure Chemical Industries, catalog number: 192-04696 )
- 1-amino-2-naphthol-4-sulfonic acid (ANSA) (NACALAI TESQUE, catalog number: 02212-12 )
- Sodium bisulfite (NaHSO3) (Wako Pure Chemical Industries, catalog number: 190-01375 )
- Sodium sulfate (Na2SO4) (Wako Pure Chemical Industries, catalog number: 192-03415 )
- Potassium dihydrogen phosphate (KH2PO4) (Wako Pure Chemical Industries, catalog number: 169-04245 )
- DTT stock solution (see Recipes)
- Protease inhibitor solution (see Recipes)
- Oligomycin solution (see Recipes)
- Homogenization buffer (see Recipes)
- 2x ATPase buffer (see Recipes)
- ATPase reaction buffer (see Recipes)
- ATP solution (see Recipes)
- Vanadate solution (see Recipes)
- Stop solution (see Recipes)
- ANSA solution (see Recipes)
- Pi standard stock solution (see Recipes)
Equipment
- Mortar (90 mm diameter) and pestle
- Refrigerated centrifuge (TOMY DIGITAL BIOLOGY, model: MX-307 )
- Ultracentrifuge (Beckman Coulter, model: OptimaTM TLX )
- Vortex (Scientific Industry, model: SI-0286 )
- Heat block (TAITEC, model: e-ThermoBucket ETB )
- Spectrophotometer (Beckman Coulter, model: DU 730 )
Note: This product has been discontinued.
Procedure
- Preparation of microsomal membranes
- Grow Arabidopsis thaliana in soil for 3 weeks at 23 °C under white light (50 µmol photons m-2 s-1) with a 16-h-light/8-h-dark cycle.
- Homogenize rosette leaves (about 100 mg) with a mortar and pestle in 2 ml ice-cold homogenization buffer and keep on ice.
- Centrifuge the homogenate at 13,000 x g for 10 min at 4 °C.
- Ultracentrifuge the supernatant at 100,000 x g for 1 h at 4 °C.
- Resuspend the pellet in 100 μl ice-cold homogenization buffer by pipetting up and down.
- Quantify the protein concentration by Bradford assay (Bradford, 1976).
- Protein concentration is adjusted to 0.45 µg/µl with homogenization buffer.
Note: Keep microsomal membranes on ice until use.
- Grow Arabidopsis thaliana in soil for 3 weeks at 23 °C under white light (50 µmol photons m-2 s-1) with a 16-h-light/8-h-dark cycle.
- Measurement of vanadate-sensitive ATP hydrolytic activity
- Mix 100 μl of microsomal membranes with 100 μl ATPase reaction buffer, split the mixture in two tubes, 90 µl each (20 µg protein), and keep on ice.
- To determine vanadate-sensitive ATPase activity, add 2 μl vanadate solution to one tube and an equal volume of 1x ATPase buffer to the other tube.
- Add 10 μl ATP solution and gently vortex.
- Incubate at 30 °C for 30 min. Gently vortex once after 15 min of incubation.
- Add 1 ml stop solution.
- Add 50 μl ANSA solution and gently vortex.
- Incubate at 24 °C for 30 min. Gently vortex once after 15 min of incubation.
- Measure absorption at 750 nm by a spectrophotometer using a cuvette.
Note: Do not centrifuge the samples.
- Mix 100 μl of microsomal membranes with 100 μl ATPase reaction buffer, split the mixture in two tubes, 90 µl each (20 µg protein), and keep on ice.
- Preparation of Pi standard curve
- Prepare Pi dilution series as shown in Table 1.
Table 1. Template for the preparation of the Pi standard curve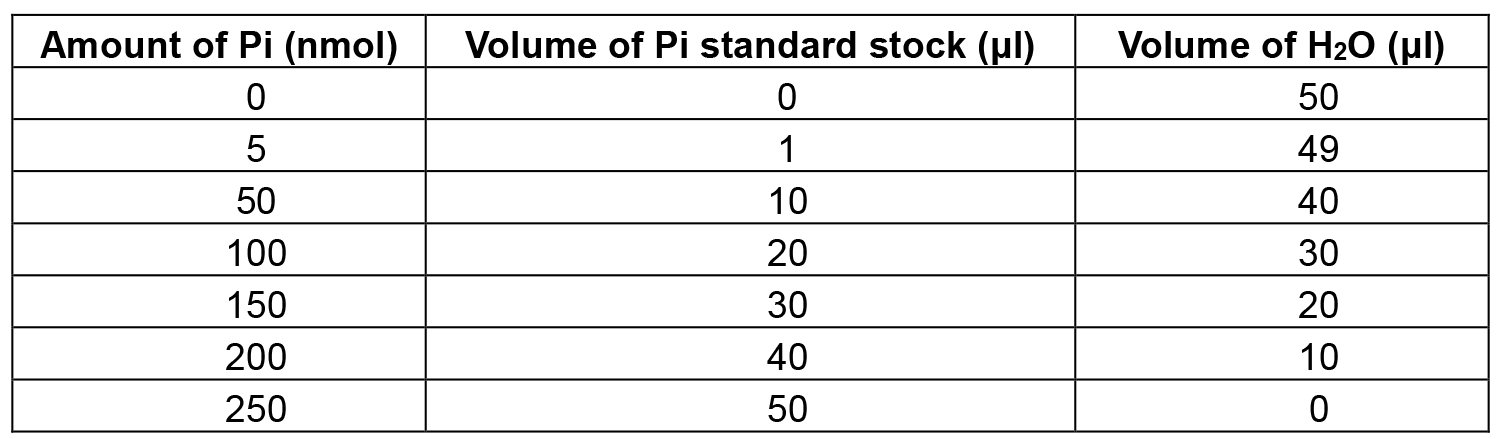
- Add 50 μl ATPase reaction buffer and 1 ml stop solution.
- Add 50 μl ANSA solution and incubate at 24 °C for 30 min. Gently vortex once after 15 min of incubation.
- Measure absorption at 750 nm using a cuvette and make a standard curve (Figure 1).
- Prepare Pi dilution series as shown in Table 1.
Data analysis
A typical Pi standard curve is shown in Figure 1. Calculate Pi content of samples using the standard curve. Vanadate-sensitive ATP hydrolytic activity is determined by subtracting Pi content in the presence of vanadate from that in the absence of vanadate, and is expressed as nmol Pi/h/mg of protein.
- Calculate a slope of Pi standard curve.
A750 = 0.0038 (Pi content [nmol]) + 0.0088 - Determine Pi content of samples from the slope.
(Pi content [nmol]) = (A750 - 0.0088)/0.0038 - Subtract Pi content in the presence of vanadate from that in the absence of vanadate.
- Divide by the amount of protein (mg), and reaction time (h). The following is an example of calculation.
Pi content in the absence of vanadate = 10 nmol
Pi content in the presence of vanadate = 5 nmol
Reaction time = 0.5 h (30 min)
Amount of protein = 0.02 mg (20 µg)
Vanadate-sensitive ATP hydrolytic activity (nmol Pi/h/mg of protein)
= ([Pi content in the absence of vanadate] - [Pi content in the presence of vanadate])/(reaction time)/(Amount of protein)
= (10 - 5)/0.5/0.02 = 500 nmol Pi/h/mg of protein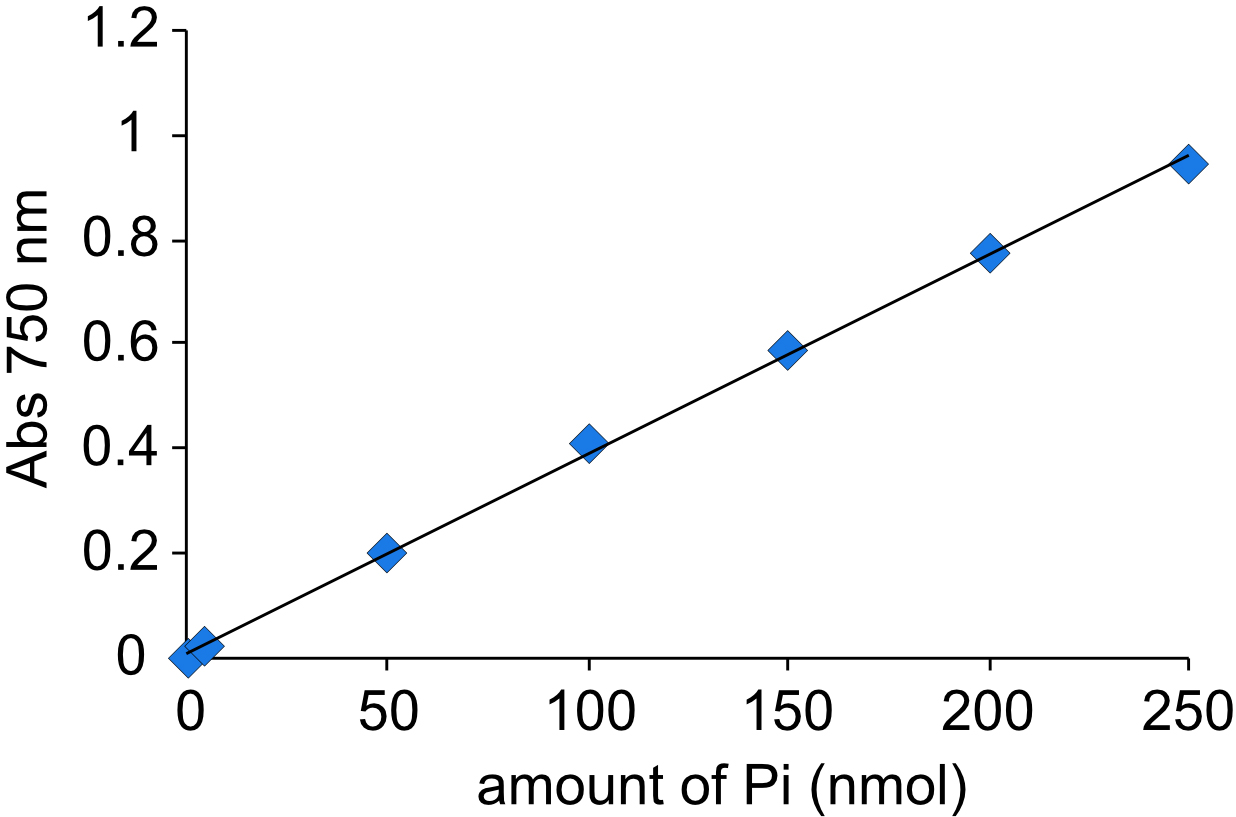
Figure 1. Standard curve generated by using known amounts of Pi. The absorption at 750 nm was measured.
Recipes
- DTT stock solution
1 M DTT in sterile water
Store at -20 °C in small aliquots - Protease inhibitor solution
200 mM PMSF and 4 mM leupeptin in DMSO
Store at 4 °C in small aliquots - Oligomycin solution
5 mg/ml oligomycin in DMSO
Store at -20 °C in small aliquots - Homogenization buffer
50 mM MOPS-KOH (pH 7.5)
100 mM NaCl
2.5 mM EDTA
10 mM NaF
5 mM DTT
1 mM PMSF
20 μM leupeptin
Store at 4 °C. Add NaF, DTT, PMSF, and leupeptin just before use - 2x ATPase buffer
60 mM Tris-MES (pH 6.5)
6 mM MgSO4
100 mM KCl
200 mM KNO3
Store at 4 °C - ATPase reaction buffer (freshly prepared)
2x ATPase buffer supplemented with:
1 mM ammonium molybdate
10 μg/ml oligomycin
0.1% (w/w) Triton X-100
0.5 mM PMSF
10 μM leupeptin - ATP solution
20 mM ATP in 1x ATPase buffer
Solution should be aliquoted in small volumes to avoid freeze-thawing and can be stored at -80 °C for at least 6 months - Vanadate solution (freshly prepared)
10 mM sodium orthovanadate in 1x ATPase buffer - Stop solution (freshly prepared)
1.3% (w/v) SDS
0.25% (w/v) sodium molybdate
0.3 N H2SO4 - ANSA solution (freshly prepared)
0.125% (w/v) 1-amino-2-naphthol-4-sulfonic acid (ANSA)
15% (w/v) NaHSO3
1% (w/v) Na2SO4 - Pi standard stock solution (freshly prepared)
5 mM KH2PO4
Acknowledgments
This protocol is adapted from Okumura et al. (2016). This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (15H05956 and 15H04386 to T.K.) and by a Grant-in-Aid for JSPS fellows (253307 to M.O.).
References
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1-2): 248-254.
- Kinoshita, T., Nishimura, M. and Shimazaki, K. (1995). Cytosolic concentration of Ca2+ regulates the plasma membrane H+-ATPase in guard cells of fava bean. Plant Cell 7(8): 1333-1342.
- Okumura, M., Inoue, S., Kuwata, K. and Kinoshita, T. (2016). Photosynthesis activates plasma membrane H+-ATPase via sugar accumulation. Plant Physiol 171(1): 580-589.
- Shimazaki, K. I. and Kondo, N. (1987). Plasma membrane H+-ATPase in guard-cell protoplasts from Vicia faba L. Plant Cell Physiol 28(5): 893-900.
- Uemura, M. and Yoshida, S. (1986). Studies on freezing injury in plant cells: II. Protein and lipid changes in the plasma membranes of Jerusalem artichoke tubers during a lethal freezing in vivo. Plant Physiol 80(1): 187-195.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Okumura, M. and Kinoshita, T. (2016). Measurement of ATP Hydrolytic Activity of Plasma Membrane H+-ATPase from Arabidopsis thaliana Leaves. Bio-protocol 6(23): e2044. DOI: 10.21769/BioProtoc.2044.
- Kinoshita, T., Nishimura, M. and Shimazaki, K. (1995). Cytosolic concentration of Ca2+ regulates the plasma membrane H+-ATPase in guard cells of fava bean. Plant Cell 7(8): 1333-1342.
Category
Plant Science > Plant physiology > Ion analysis
Plant Science > Plant biochemistry > Protein > Activity
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









