- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Microplate Assay to Study Carboxypeptidase A Inhibition in Andean Potatoes
(*contributed equally to this work) Published: Vol 6, Iss 23, Dec 5, 2016 DOI: 10.21769/BioProtoc.2032 Views: 10228
Reviewed by: Arsalan DaudiManjula MummadisettiDaniel F. Caddell

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

A Semi-throughput Procedure for Assaying Plant NADP-malate Dehydrogenase Activity Using a Plate Reader
Kevin Baudry and Emmanuelle Issakidis-Bourguet
Aug 20, 2023 1474 Views

An in vitro Assay to Probe the Formation of Biomolecular Condensates
Yu Zhang and Shen Lisha
Sep 5, 2023 3209 Views

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1824 Views
Abstract
Metallocarboxypeptidases (MCP) are zinc-dependent exopeptidases that catalyze the hydrolysis of C-terminal amide bonds in proteins and peptides. They are involved in a wide range of physiological processes and have recently emerged as relevant drug targets in biomedicine (Arolas et al., 2007). In this context, the study and discovery of new MCP inhibitors from plants constitute a valuable approach for the development of new therapeutic strategies. Herein we describe a simple and accessible microplate method for the study of the specific and dose-response carboxypeptidase A inhibitory activities present in Andean potato tubers. Our protocol combines an extraction method optimized for small protein inhibitors in plant tissues, with the measurement of enzyme kinetics using a microplate reader. These instruments are capable of reading small sample volumes, for many samples in a very short time-frame, therefore reducing the time and costs of high-throughput screening experiments. Although this protocol describes the study of Andean potatoes, our approach is also applicable to the analysis other plant samples.
Keywords: MetallocarboxypeptidaseBackground
In higher plants, small proteinaceous protease inhibitors are wound-induced molecules produced as a part of its defense system against insect attack (Graham et al., 1981; Villanueva et al., 1998). Among the studied inhibitors, only two are specific for MCP, i.e., the potato carboxypeptidase inhibitor (PCI) and its close homolog found in tomato plants (TCI). Over the last few decades, the presence of MCP inhibitors in Solanaceae has been extensively reported, revealing potato (Solanum tuberosum) as one of the most important sources of MCP inhibitors (Hass et al., 1979; Obregón et al., 2012; Lufrano et al., 2015). In humans, MCP action is exquisitely regulated and dysregulation of its function might lead to disease or even to cell death (Arolas et al., 2007). In fact, MCP have been associated with human pathologies such as acute pancreatitis (Appelros et al., 1998), diabetes (Cool et al., 1997), several types of cancer (Ross et al., 2009; Sun et al., 2016; Abdelmagid et al., 2008; Tsakiris et al., 2008), fibrinolysis (Valnickova et al., 2007), inflammation (Deiteren et al., 2009) or neurodegeneration (Rogowski et al., 2010). In this context, there is an interest in the discovery of new MCP inhibitors, and thus we focus our studies in potatoes that are native from the Andean region of South America. In this region, thousands of different potato varieties coexist, constituting a natural reservoir for the discovery of novel MCP inhibitors (Figure 1).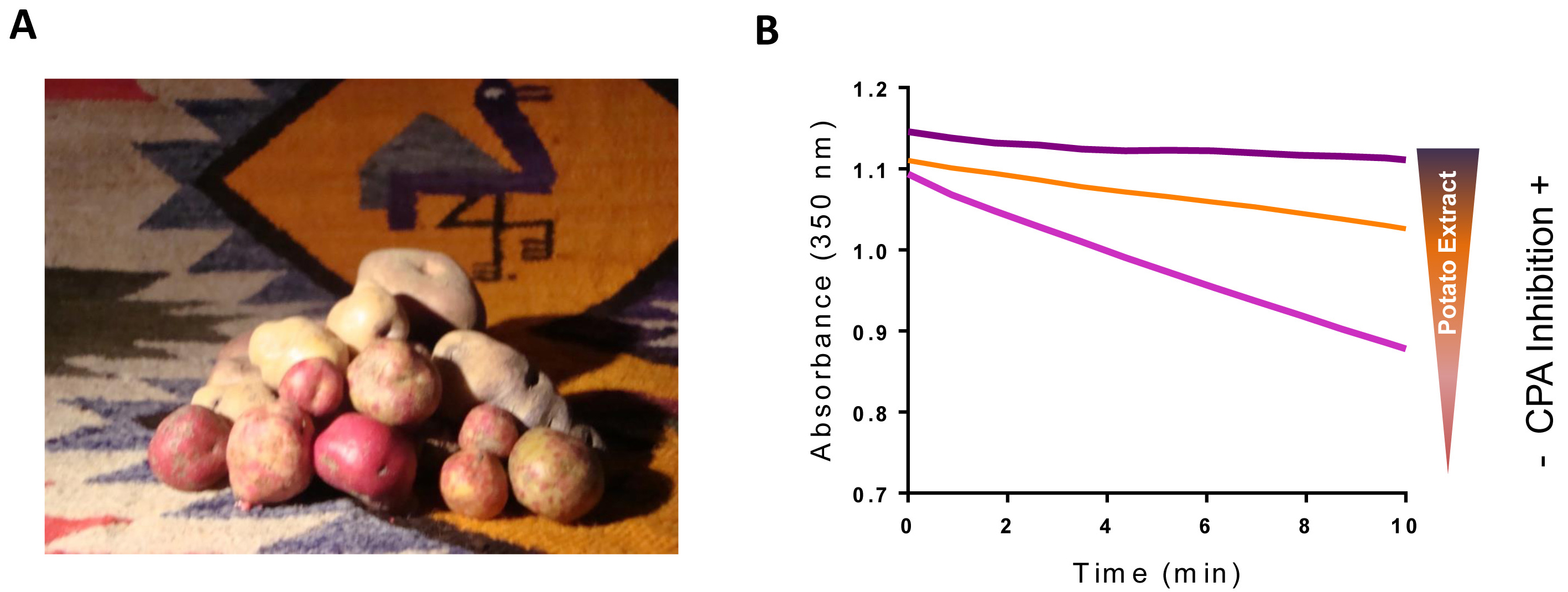
Figure 1. Andean potatoes and potato extract CPA inhibitory activity. A. Picture displaying the large number of potato varieties found within the Andean region. Currently in this region coexist thousands of Andean varieties of Solanum tuberosum (Machida-Hirano, 2015; Clausen et al., 2010). B. Effects of potato extracts on bCPA activity. The activity of bovine CPA (bCPA) was measured using the substrate N-(4-methoxyphenylazoformyl)-Phe-OH determining the decrease in absorbance at 340 nm in function of the time. Due to its high content in MCP inhibitors, the addition of potato extract to the reaction decreases the rate of substrate hydrolysis in a dose-response manner.
Here, we describe a simple protocol to determine the specific and dose-response carboxypeptidase A inhibitory activity present in Andean potatoes and in other biological extracts using microplates. The major advantage of this protocol over other available approaches (Yanes et al., 2007) is that the use of microplates allows multiple enzymatic measurements to be done in a single experiment, therefore reducing the time and costs.
Materials and Reagents
- 50 ml tubes
- 96-well microplates, clear flat bottom (Corning, catalog number: 3364 )
- Syringe filters, 0.45 μm pore size (EMD Millipore, catalog number: SLHP033RS )
- Potato tubers
- General laboratory materials and instrumentation (e.g., micropipettes, microtubes, tips)
- Bradford assay kit, e.g., Coomassie Plus Assay Kit (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 23236 )
- Bovine serum albumin (BSA)
- N-(4-methoxyphenylazoformyl)-Phe-OH·potassium salt (Bachem, catalog number: M-2245 )
- Trizma® base (Sigma-Aldrich, catalog number: T1503 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 258148 )
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: D4540 )
- Bovine carboxypeptidase A (bCPA) (Sigma-Aldrich, catalog number: C9268 )
- Carboxypeptidase A reaction buffer/Extraction buffer (see Recipes)
- 2 mg/ml bCPA stock solution (see Recipes)
- 10x bCPA working solution (see Recipes)
- 1,000x substrate stock solution (see Recipes)
- 10x substrate working solution (see Recipes)
Equipment
- Laboratory blender or equivalent (Oster, catalog number: 004093-008-NP0 )
- Refrigerated centrifuge (suitable for volumes of 50 ml) (Beckman Coulter, model: Avanti J-26 XPI )
- UV-Vis microplate spectrophotometer system capable of operating at 340 and 595 nm (e.g., PerkinElmer, model: Victor X 2030-0050 or other equivalent spectrophotometer)
- pH meter (HACH LANGE SPAIN, Crison, model: GLP 21 )
- 37 °C oven (e.g., Thermo Fisher Scientific, Thermo ScientificTM, model: Heratherm Compact Microbiological Incubator )
- Multichannel pipette (e.g., Technology Networks, model: CappAero Multichannel Pippete 25-200 μl )
Software
- GraphPad Prism 5 software (GraphPad Software, Ing USA)
Procedure
- Preparation of Andean potato extracts and protein quantification
- Wash two mature potato tubers (stage five, according to Johnson, 2008) with distilled water. Peel, weigh (in our case 11.50 g) and dice them into small pieces of about 2 x 2 cm in size (see Figures 2A and 2B).
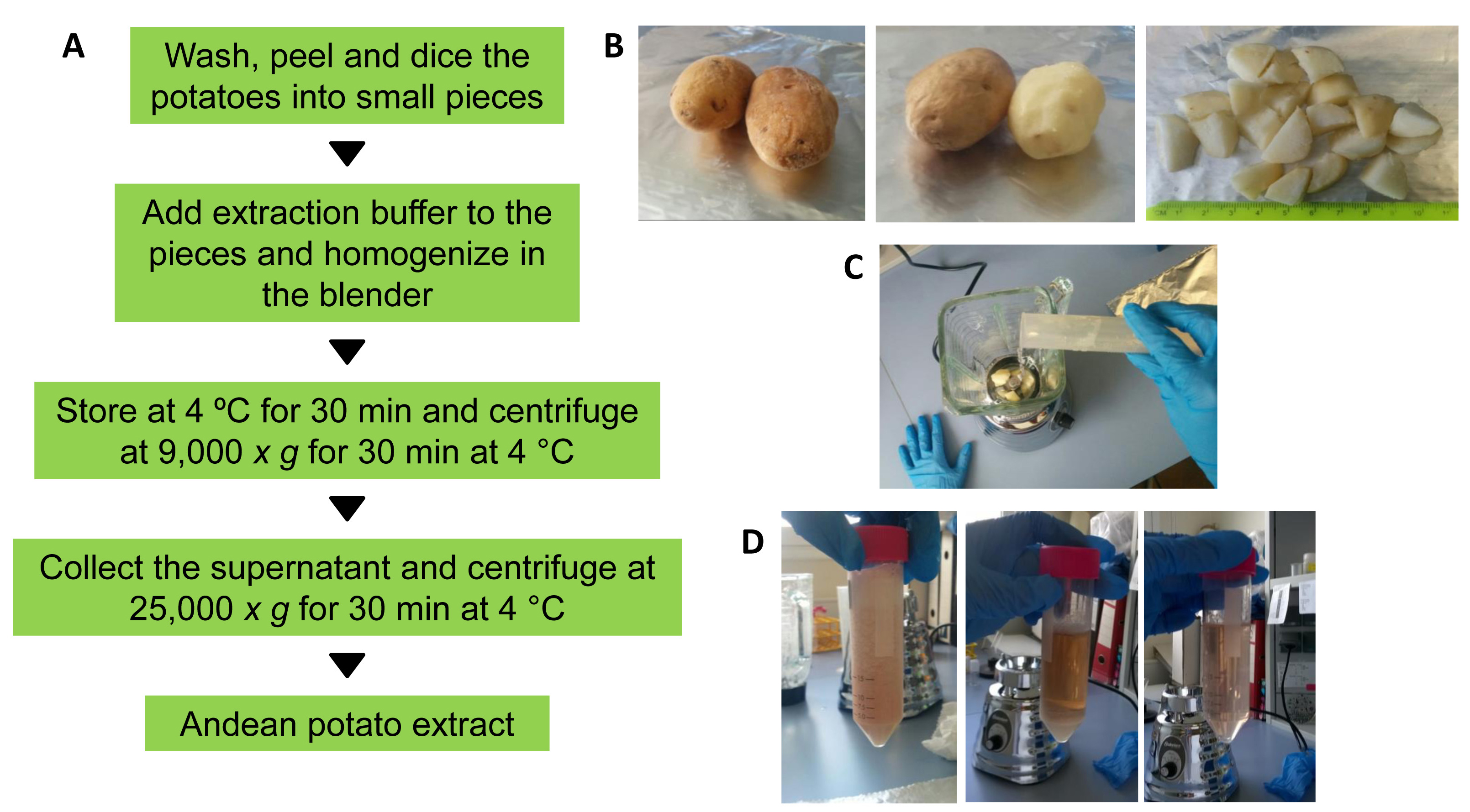
Figure 2. Preparation of potato extracts. A. Workflow followed to obtain the crude potato extract enriched in MCP inhibitors. B. Potato peeling and dicing. C. Mixing of the diced potatoes with the extraction buffer into the blender. D. The appearance of the potato extract before and after two centrifugation steps (from left to right). - Mix the potato pieces with three volumes of ice-cold extraction buffer (in our case 34.50 ml; a mass/volume ratio of 1:3) into the cooled blender and homogenize at gentle speed for 5 min. (Figures 2A and 2C).
Note: The homogenization should be performed in several sessions of 15-30 sec each, with 60 sec intervals between sessions to prevent excessive heating of the sample. - After homogenization, transfer the potato homogenate to 50 ml tubes and centrifuge the sample at 9,000 x g for 30 min at 4 °C.
- Transfer the supernatant to a clean 50 ml centrifuge tube and spin at 25,000 x g for 30 min at 4 °C (Figures 2A and 2D). After the second centrifugation step, collect the resultant supernatant and filter through a 0.45 μm syringe filter to eliminate protein aggregates. The resultant potato extract can be stored at -20 °C until analyzed.
- Determine the protein concentration of the samples using the Coomassie Plus Assay Kit, according to the manufacturer’s instructions. In brief, prepare a final volume of 500 μl of each of the six standard solutions containing 0, 2, 5, 10, 15 and 20 μg/ml of BSA and appropriate dilutions of the sample/s. Transfer 150 μl of each standard and potato extract samples into different wells of a 96-well microplate. BSA standards and potato extract samples should be assayed in triplicate. Then, add 150 μl of the Coomassie Plus reagent (see Materials and Reagents section) to each well and mix using the micropipette, by pipetting up and down carefully. After 5 min incubation at room temperature, measure the absorbance at 595 nm using a UV-Vis microplate spectrophotometer. Typically, we obtained 1-2 mg/ml of protein in the extracts.
- Wash two mature potato tubers (stage five, according to Johnson, 2008) with distilled water. Peel, weigh (in our case 11.50 g) and dice them into small pieces of about 2 x 2 cm in size (see Figures 2A and 2B).
- Enzymatic assays
- Determination of bCPA specific inhibitory activity
- Prepare triplicates of the reaction mixtures (Table 1) in a microplate without adding the substrate. Calculate the volume of potato extract to be added, in order to obtain 20% to 80% of bCPA inhibition. To obtain these inhibition levels, we typically add around 5-30 μg/ml of final protein concentration to the assay.
Table 1. Reaction mixture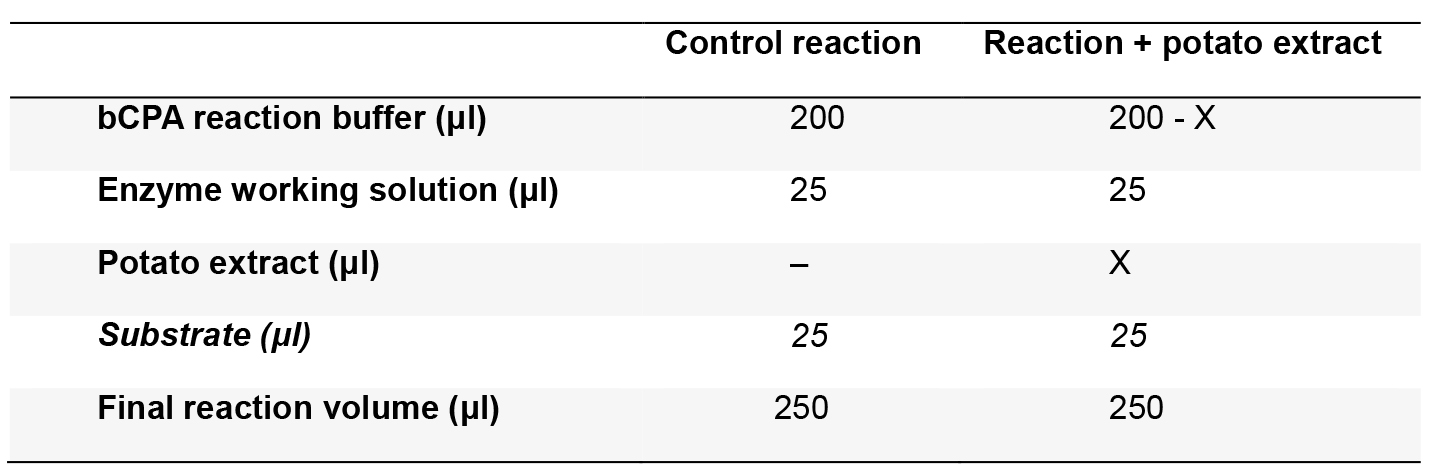
Where X is the volume (in μl) of potato extract to be assayed. The substrate should be added immediately before plate reading (see step B1c below) - Cover the microplate with a lid and incubate at 37 °C for 15 min.
- Add 25 μl of substrate working solution to each well, mix carefully and thoroughly, by pipetting up and down carefully with a multichannel micropipette (A graphical demonstration of steps B1a, B1b and B1c is shown in Video 1).
Note: the homogenization should be performed in no longer than 60 sec, to avoid significant consumption of the substrate before absorbance monitoring. - Perform absorbance measurements at 340 nm every 30 sec for 10 min.
- One unit of inhibitory activity is defined as the amount of inhibitor able to reduce one unit of bCPA activity, which in turn corresponds to the amount of enzyme that hydrolyzes 1.0 μmol of N-(4-methoxyphenylazoformyl)-Phe-OH per min at 25 °C. Consequently, Equation 1 can be used to calculate the Specific Inhibitory Activity (SIA) found in potato extracts.
Equation 1: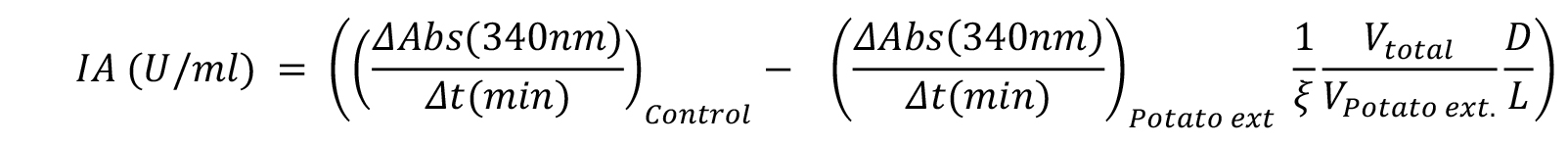
Where,
IA is the Inhibitory Activity in U/ml,
ΔAbs/Δt is the absorbance variation per unit of time (in min) during the reaction in absence (control), and in presence of the potato extract (Potato ext), respectively,
ξ is the extinction coefficient for the substrate N-(4-methoxyphenylazoformyl)-Phe-OH (ξ 350 nm = 19 [μmoles/ml]-1 cm-1),
Vtotal is the assay volume and VPotato ext is the volume of extract added to the reaction,
D and L are the dilution factor for the extract and the path length (in cm), respectively. Typically, the path length in a microplate for a volume of 250 μl is 0.7 cm. However, for different reaction volumes, or for a more accurate calculation, check your 96-well microplate manufacturer instructions.
Then, calculate directly the SIA by dividing the resultant IA value by the protein concentration of the sample in mg/ml (see Equation 2 and examples in Table 2).
Equation 2: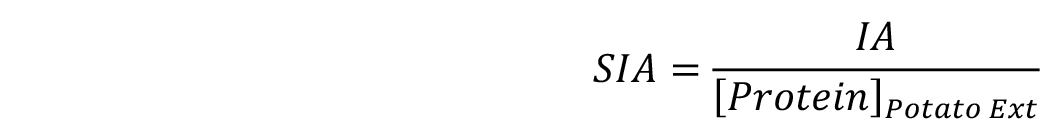
- Prepare triplicates of the reaction mixtures (Table 1) in a microplate without adding the substrate. Calculate the volume of potato extract to be added, in order to obtain 20% to 80% of bCPA inhibition. To obtain these inhibition levels, we typically add around 5-30 μg/ml of final protein concentration to the assay.
- Determination of the IC50: Dose-Response curve assay (see Figure 3)
- Prepare the same control reaction as described in step B1a and prepare at least 12 additional reaction mixtures with different final concentrations of the potato extract in the assay, ranging from 0 to 300 μg/ml, (or even with higher concentration to obtain the complete inhibition of bCPA activity).
Note: We typically assay a total of 15 different extract final concentrations, containing 0, 1, 2, 3, 5, 10, 15, 20, 30, 75, 100, 150, 200, 250 and 300 μg/ml of protein.
Fit the results obtained to the following Equation 3 and determine the IC50 value.
Equation 3:
Where,
X is the log-transformed protein concentration assayed,
Y is the normalized bCPA activity (relative to the control condition and expressed as a percentage of the maximal activity).
Y values can be calculated for each extract concentration, using the following Equation 4.
Equation 4: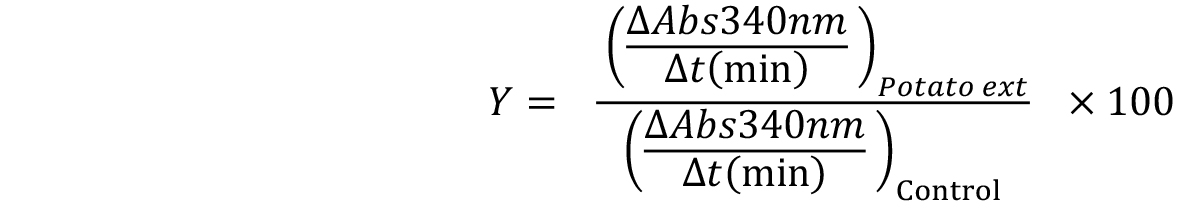
The IC50 value is the extract concentration necessary to reach a 50% of bCPA inhibition (Copeland, 2005). See representative examples of dose-response bCPA inhibitory plots in Figure 3 and the corresponding IC50 values in Table 2.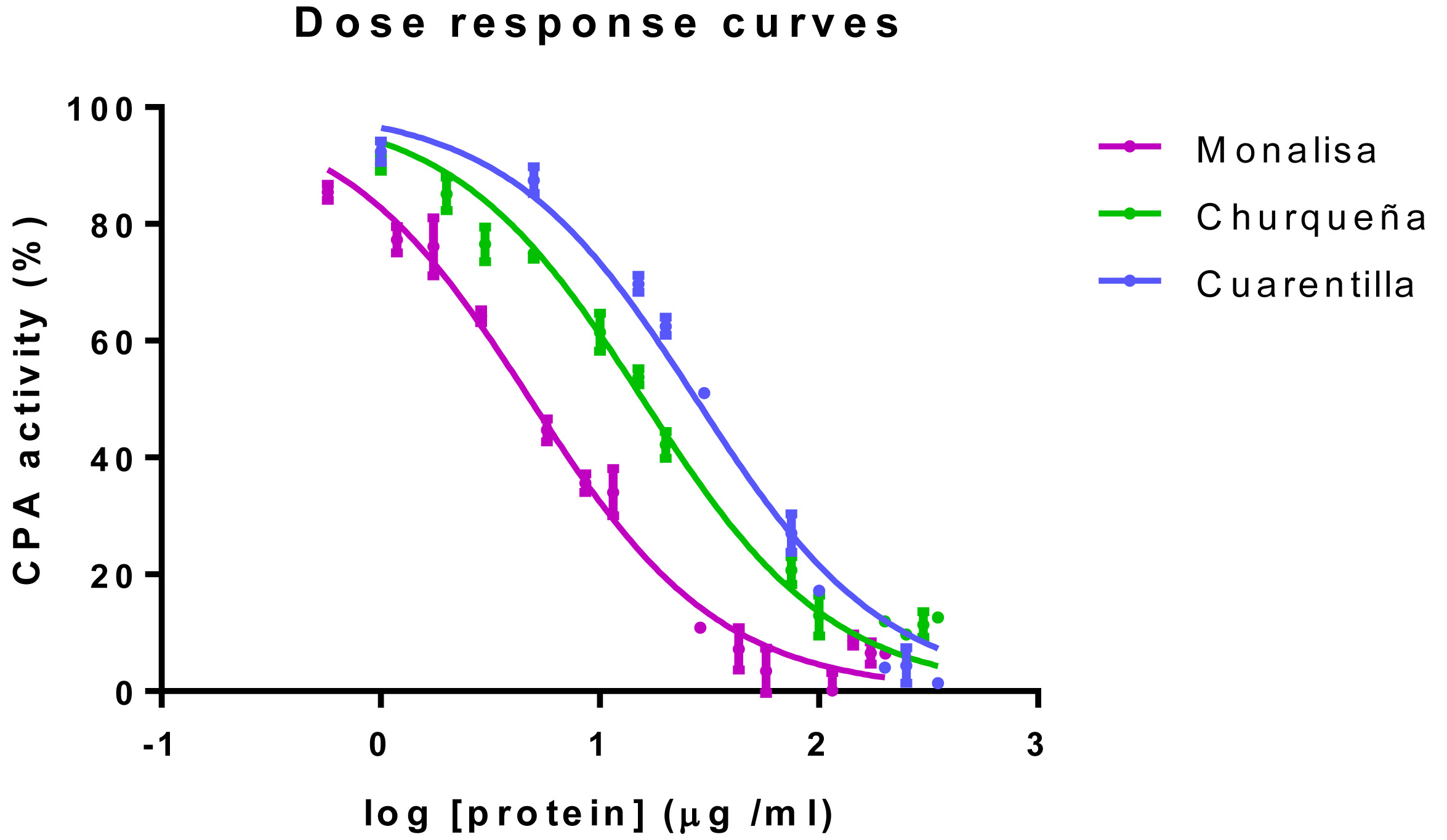
Figure 3. Examples of dose-response inhibitory plots. Representative examples of dose-response inhibitory curves determined for three different varieties of potatoes; Solanum tuberosum subsp. tuberosum var. monalisa (Monalisa, magenta solid line), Solanum tuberosum subsp. andigenum var. churqueña (Churqueña, green solid line), Solanum tuberosum subsp. andigenum var. cuarentilla; (Cuarentilla, blue solid line).
Table 2. Summary of the inhibitory activities found in different varieties of potatoes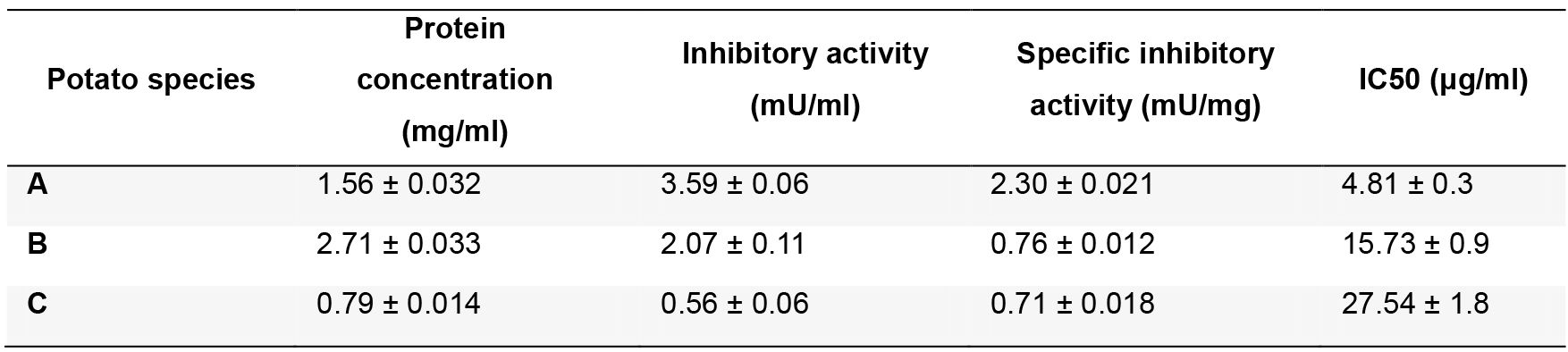
A: Solanum tuberosum subsp. tuberosum var. monalisa; B: Solanum tuberosum subsp. andigenum var. churqueña; C: Solanum tuberosum subsp. andigenum var. cuarentilla
The two protocols for the determination of SIA and IC50 are summarized in the scheme of Figure 4 and Video 1.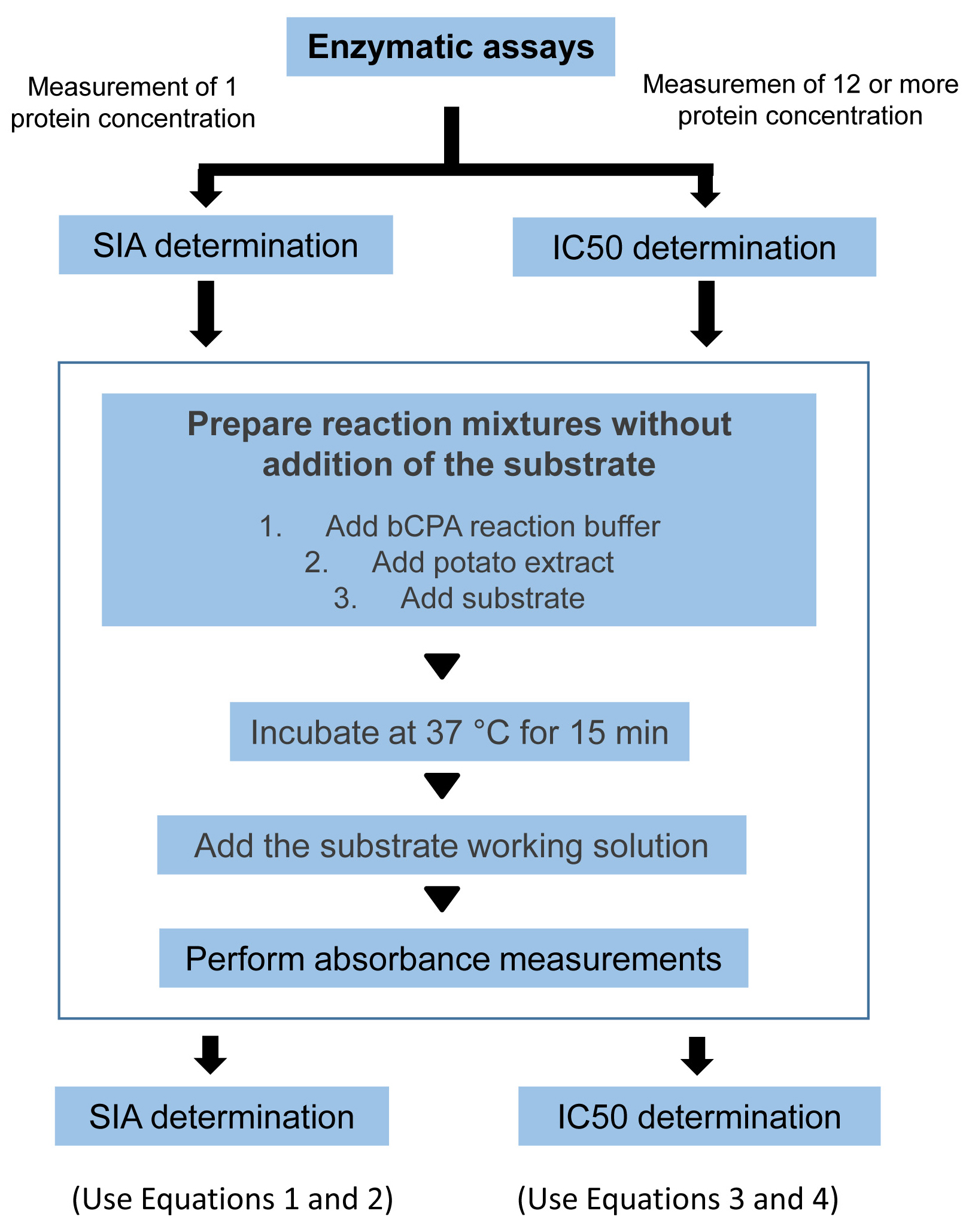
Figure 4. Enzymatic assays. General workflow for the measurement of inhibitory activity and determination of the SIA and IC50.Video 1. Video demonstration of the protocol to perform the enzymatic measurements
- Prepare the same control reaction as described in step B1a and prepare at least 12 additional reaction mixtures with different final concentrations of the potato extract in the assay, ranging from 0 to 300 μg/ml, (or even with higher concentration to obtain the complete inhibition of bCPA activity).
- Determination of bCPA specific inhibitory activity
Data analysis
Data fittings and IC50 determination were performed using GraphPad Prism 5 software (GraphPad Software, Ing USA) (Motulsky et. al., 2007). Only fittings with an R2 > 0.97 were considered.
Notes
- Freshly collected samples display higher inhibitory activities; however, frozen samples can also be used.
- The protocol described here is suitable for any scale of sample preparation, therefore volumes and reagent quantities should be scaled proportionally.
- Here we used a laboratory blender that allows a complete sample blending and homogenization. Alternatively, other appropriate homogenization systems can be used.
- The soluble fraction resultant from the step A4 is a crude extract rich in MCP inhibitors. Other proteinaceous and chemical compounds soluble in the homogenization buffer can be also present in the sample. For a higher MCP inhibitor enrichment (e.g., for Ki determination), further purification procedures could be addressed (Pearce and Ryan, 1983).
- We used a commercial Bradford assay kit; however, non-commercial Bradford reagents or other different quantification assays can be used (Bradford, 1976; He, 2011a and 2011b).
- The microplate reactions can alternatively be performed in conventional spectrophotometer cuvettes by scaling up the reagents proportionately.
- Mix the enzyme suspension thoroughly to ensure complete mixing. It is strongly recommended to follow manufacturer’s instructions for an accurate enzyme preparation.
- The substrate concentration used is such that the enzyme is at the maximum velocity. The enzyme concentration might be adjusted to consume less than 5-10% of substrate.
Recipes
- Carboxypeptidase A reaction buffer/Extraction buffer (1 L, 20 mM Tris-HCl, 500 mM NaCl, pH 7.5)
2.42 g of Trizma base
29.22 g of NaCl
MilliQ water up to 900 ml
Adjust solution to pH 7.5 by addition of 5 N HCl
Adjust the final volume with MilliQ water to 1,000 ml
Store at 4 °C up to two weeks - 2 mg/ml bCPA stock solution
Prepare 1 ml of a 2 mg/ml bCPA solution (~57 µM) in reaction buffer
Note: We typically dilute 100 µl of commercial bCPA with 900 µl of carboxypeptidase A reaction buffer. Note that different batches of bCPA may have different enzyme concentrations, therefore the dilutions should be adjusted accordingly.
Divide into 20 μl aliquots
For short-term storage, store at 4 °C; for long-term storage, store at -20 °C - 10x bCPA working solution
Prepare 10 ml of a 50 nM enzyme solution by diluting 8.77 μl of bCPA stock solution in 9.991 ml of carboxypeptidase A reaction buffer
For short-term storage, store at 4 °C; for long-term storage, store at -20 °C - 1,000x substrate stock solution (100 mM N-[4-methoxyphenylazoformyl]-Phe-OH)
Dissolve 100 mg of N-(4-methoxyphenylazoformyl)-Phe-OH·potassium salt (Mock et al., 1996) in 2.74 ml of DMSO
Divide into 100 μl aliquots and store at -20 °C - 10x substrate working solution, 1 mM (N-[4-methoxyphenylazoformyl]-Phe-OH)
Dilute 100 μl of the substrate stock solution to a final volume of 10 ml with carboxypeptidase A reaction buffer
Store at -20 °C
Acknowledgments
The protocol described here was adapted from previously published studies (Covaleda et al., 2012; Lufrano et al., 2015; Sanglas et al., 2009). This work was supported by the Spanish Ministry of Innovation and Competitiveness grant BIO2013-44973-R and by UNLP, Argentina grant PPID/X014. The authors acknowledge support of the CONICET (PIP 0120), Universidad Nacional de La Plata (UNLP), PPID X/014, Bilateral Cooperation Program MinCyT-MICINN (project ES/09/24-AR2009/006) and Proyectos Redes Universitarias, PPUA, SPU, Argentina.
References
- Abdelmagid, S. A. and Too, C. K. (2008). Prolactin and estrogen up-regulate carboxypeptidase-d to promote nitric oxide production and survival of MCF-7 breast cancer cells. Endocrinology 149(10): 4821-8.
- Appelros, S., Thim, L. and Borgstrom, A. (1998). Activation peptide of carboxypeptidase B in serum and urine in acute pancreatitis. Gut 42(1): 97-102.
- Arolas, J. L., Vendrell, J., Aviles, F. X. and Fricker, L. D. (2007). Metallocarboxypeptidases: emerging drug targets in biomedicine. Curr Pharm Des 13(4): 349-366.
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Clausen, A. M., Ispizúa, N. and Digilio. (2010). Native andean potato varieties in argentina: conservation and evaluation of an endangered genetic resource contents. AmJPSB 3(1): 72-82.
- Cool D. R., Normant E., Shen F., Chen H. C., Pannell L., Zhang Y. and Loh Y. P. (1997). Carboxypeptidase E is a regulated secretory pathway sorting receptor: genetic obliteration leads to endocrine disorders in Cpefat mice. Cell 88(1): 73–83.
- Copeland R. (2005). Lead optimization and SAR for reversible inhibitors. In: Copeland R. (Ed). Evaluation of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists. John Wiley & Sons, pp: 125-128.
- Covaleda, G., del Rivero, M. A., Chavez, M. A., Aviles, F. X. and Reverter, D. (2012). Crystal structure of novel metallocarboxypeptidase inhibitor from marine mollusk Nerita versicolor in complex with human carboxypeptidase A4. J Biol Chem 287(12): 9250-9258.
- Deiteren, K., Hendriks, D., Scharpe, S. and Lambeir, A. M. (2009). Carboxypeptidase M: Multiple alliances and unknown partners. Clin Chim Acta 399(1-2): 24-39.
- Graham, J. S. and Ryan, C. A. (1981). Accumulation of a metallo-carboxypeptidase inhibitor in leaves of wounded potato plants. Biochem Biophys Res Commun 101(4): 1164-1170.
- Hass, G. M. and Derr, J. E. (1979). Purification and characterization of the carboxypeptidase isoinhibitors from potatoes. Plant Physiol 64(6): 1022-1028.
- He, F. (2011a). Bradford protein assay. Bio-protocol Bio101: e45.
- He, F. (2011b). BCA (bicinchoninic acid) protein assay. Bio-protocol Bio101: e44.
- Johnson, D. A. (2008). Potato Health Management (2nd ed.). American Phytopathology Society.
- Lufrano, D., Cotabarren, J., Garcia-Pardo, J., Fernandez-Alvarez, R., Tort, O., Tanco, S., Aviles, F. X., Lorenzo, J. and Obregon, W. D. (2015). Biochemical characterization of a novel carboxypeptidase inhibitor from a variety of Andean potatoes. Phytochemistry 120: 36-45.
- Machida-Hirano, R. (2015). Diversity of potato genetic resources. Breed Sci 65(1): 26-40.
- Mock, W. L., Liu,Y. and Stanford, D. J. (1996). Arazoformyl peptide surrogates as spectrophotometric kinetic assay substrates for carboxypeptidase A. Anal Biochem 239(2): 218-222.
- Motulsky, H. (2007). Prism 5 statistics guide. GraphPad Software.
- Obregón, W. D.; Ghiano, N; Tellechea, M., Cisneros, S., Lazza, C. M.; López, L.M.I. and F. X. Avilés (2012). Detection and characterization of a new metallocarboxypeptidase inhibitor from Solanum tuberosum cv. Desirèe using proteomic techniques. Food Chem 133(4): 1063–1068.
- Pearce, G. and Ryan, C. A. (1983). A rapid, large-scale method for purification of the metallo-carboxypeptidase inhibitor from potato tubers. Anal Biochem 130(1): 223-225.
- Rogowski, K., van Dijk, J., Magiera, M.M., Bosc, C., Deloulme, J.C., Bosson, A., Peris, L., Gold, N.D., Lacroix, B., Bosch Grau, M., Bec, N., Larroque, C., Desagher, S., Holzer, M., Andrieux, A., Moutin, M.J. and Janke, C. (2010). A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell 143(4): 564–578.
- Ross P. L., Cheng I., Liu X., Cicek M. S., Carroll P. R., Casey G. and Witte J. S. (2009). Carboxypeptidase 4 gene variants and early-onset intermediate-to-high risk prostate cancer. BMC Cancer 9: 69.
- Sanglas, L., Aviles, F. X., Huber, R., Gomis-Ruth, F. X. and Arolas, J. L. (2009). Mammalian metallopeptidase inhibition at the defense barrier of Ascaris parasite. Proc Natl Acad Sci U S A 106(6): 1743-1747.
- Sun, L., Wang, Y., Yuan, H., Burnett, J., Pan, J., Yang, Z., Ran, Y., Myers, I. and Sun, D. (2016). CPA4 is a novel diagnostic and prognostic marker for human Non-Small-Cell lung cancer. J Cancer 7(10):1197-204.
- Tsakiris I., Soos G., Nemes Z., Kiss S. S., Andras C., Szanto J. and Dezso B. (2008). The presence of carboxypeptidase-M in tumour cells signifies epidermal growth factor receptor expression in lung adenocarcinomas: the coexistence predicts a poor prognosis regardless of EGFR levels. J Cancer Res Clin Oncol 134(4): 439-451.
- Valnickova, Z., Thogersen, I. B., Potempa, J. and Enghild, J. J. (2007). Thrombin-activable fibrinolysis inhibitor (TAFI) zymogen is an active carboxypeptidase. J Biol Chem 282(5): 3066-3076.
- Villanueva, J., Canals, F., Prat, S., Ludevid, D., Querol, E. and Aviles, F. X. (1998). Characterization of the wound-induced metallocarboxypeptidase inhibitor from potato cDNA sequence, induction of gene expression, subcellular immunolocalization and potential roles of the C-terminal propeptide. FEBS Lett 440(1-2): 175-182.
- Yanes, O., Villanueva, J., Querol, E. and Aviles, F. (2007). Enzymatic measurements for the detection of trypsin and carboxypeptidase A inhibitory activity. Protocol Exchange.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Tellechea, M. E., Garcia-Pardo, J., Cotabarren, J., Lufrano, D., Bakas, L., Avilés, F. X., Obregon, W. D., Lorenzo, J. and Tanco, S. (2016). Microplate Assay to Study Carboxypeptidase A Inhibition in Andean Potatoes. Bio-protocol 6(23): e2032. DOI: 10.21769/BioProtoc.2032.
Category
Plant Science > Plant biochemistry > Protein > Activity
Plant Science > Plant molecular biology > Protein
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link















