- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Evaluation of Cross-presentation in Bone Marrow-derived Dendritic Cells in vitro and Splenic Dendritic Cells ex vivo Using Antigen-coated Beads
Published: Vol 6, Iss 22, Nov 20, 2016 DOI: 10.21769/BioProtoc.2015 Views: 12804
Reviewed by: Ivan ZanoniPer AndersonShanie Saghafian-Hedengren

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation and Ex Vivo Testing of CD8+ T-Cell Division and Activation Using Mouse Splenocytes
Melissa Dolan [...] John M.L. Ebos
Aug 20, 2025 3878 Views

Detection of Autophagy in Human Peripheral Blood Mononuclear Cells Using Guava® Autophagy and Flow Cytometry
Melanie Scherer [...] Jörg Bergemann
Sep 20, 2025 1417 Views

Protocol for the Isolation and Analysis of Extracellular Vesicles From Peripheral Blood: Red Cell, Endothelial, and Platelet-Derived Extracellular Vesicles
Bhawani Yasassri Alvitigala [...] Lallindra Viranjan Gooneratne
Nov 5, 2025 1448 Views
Abstract
Antigen presentation by MHC class I molecules, also referred to as cross-presentation, elicits cytotoxic immune responses. In particular, dendritic cells (DC) are the most proficient cross-presenting cells, since they have developed unique means to control phagocytic and degradative pathways.
This protocol allows the evaluation of antigen cross-presentation both in vitro (by using bone marrow-derived DC) and ex vivo (by purifying CD8+ DC from spleen after incorporation of particulate antigen) using ovalbumin (OVA)-coupled particles. Cross-presentation efficiency is measured by three different readouts: the B3Z hybridoma T cell line (Karttunen et al., 1992) and stimulation of antigen-specific CD8+ T cells (OT-I) (Kurts et al., 1996), either analyzing OT-I activation by CD69 expression or OT-I proliferation after labeling them with carboxyfluorescein succinimidyl ester (CFSE). By using this approach, we could show recently that DCs are able to increase cross-presentation efficiency transiently upon engagement of TLR4 (Alloatti et al., 2015).
Background
In mouse, antigen-presenting cells (APC) are able to take up exogenous antigens to process them and to load peptides derived from such exogenous antigens onto major histocompatibility complex (MHC) class I molecules. Peptide-MHC I complexes are subsequently transported to the plasma membrane, where they might be presented to CD8+ T cells thereby promoting T cell activation, a process referred to as cross-presentation (Joffre et al., 2012). Among the different APC, dendritic cells (DC) excel at cross-presentation and comprise of different subpopulations expressing the XCR1 marker, which have been shown to cross-present antigens very efficiently (i.e., CD8+ resident DC from spleen and CD103+ migratory DC from skin and lung) (Dorner et al., 2009; Crozat et al., 2011). While the purification of DC residing in spleen or migratory DC is feasible, it is laborious and expensive. In order to study the cell biology of DC, primary cultures of bone marrow-derived DC (BMDC) can be easily differentiated from myeloid progenitors by culturing them with GM-CSF. Even though BMDC cannot be associated with any particular DC subtype (perhaps inflammatory DC), they constitute a valuable tool to study the main characteristics of DC cell biology. Herein, we introduce a detailed protocol to analyze cross-presentation of particulate antigen by BMDC, but also by CD8+ splenic DC. Although previous protocols included different antigen forms and read-outs, the protocol described here aims to analyze cross-presentation in a comprehensive and concise way in different DC types.
Materials and Reagents
- 2 ml Eppendorf tubes
- 15 ml centrifuge tubes
- 50 ml centrifuge tubes
- 14 ml tubes
- FisherbrandTM cell strainer (Thermo Fisher Scientific, Fisher Scientific, catalog number: 22-363-548 )
- Pre-separation filters (30 μm) (Miltenyi Biotec, catalog number: 130-041-407 )
- 1 ml insulin syringes (Terumo Medical, catalog number: SS+01H1 )
- 2.5 ml syringes
- 25 G needles (Terumo, catalog number: AN*2516R1 )
- Non-treated 96-well plates (Coring, Falcon®, round bottom, catalog number: 351177 )
- Non-treated 6-well plates (Sigma-Aldrich, catalog number: M8562-100EA )
- Non-treated Petri dish, 145 x 20 mm (Greiner Bio One, catalog number: 639161 )
- Mice: C57BL/6 and C57BL/6 recombination activating gene 1-deficient OT-I TCR (Vα2, Vβ5) transgenic mice were obtained from Charles River Laboratories (CDTA, Orleans, France)
- B3Z T cell line (a Kb-restricted, OVA-specific CD8+ T cell hybridoma) (Kurts et al., 1996)
- Low endotoxin OVA (50 mg/ml stock) (Worthington Biochemical, catalog number: LS003062 )
- OVA peptide 257-264 (SIINFEKL) (Polypeptide, catalog number: SC1302 )
- Polybead® polystyrene 3.0 micron microspheres (Polysciences, catalog number: 17134 )
- Polybead® dyed blue 1.0 micron microspheres (Polysciences, catalog number: 15712 )
- PBS (1x, pH 7.4) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010-023 )
- Glycine (Sigma-Aldrich, catalog number: 50046 )
- Glutaraldehyde (25%) (Sigma-Aldrich, catalog number: G5882 )
- Iscove’s modified Dulbecco’s medium (IMDM) (Sigma-Aldrich, catalog number: I3390-500ML )
- Penicillin-streptomycin (10,000 U/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- RPMI with GlutaMAXTM (Thermo Fisher Scientific, catalog number: 61870-010 )
- GlutaMAXTM supplement (100x) (Thermo Fisher Scientific, GibcoTM, catalog number: 35050061 )
- β-mercaptoethanol (Thermo Fisher Scientific, GibcoTM, catalog number: 21985-023 )
- MEM non-essential amino acids solution (100x) (Thermo Fisher Scientific, GibcoTM, catalog number: 11140050 )
- Sodium pyruvate (100 mM) (Thermo Fisher Scientific, GibcoTM, catalog number: 11360070 )
- CPRG (Roche Diagnostics, catalog number: 10884308001 )
- Fixable viability dye eFluor 780 (dilution 1/10,000) (Affymetrix, eBioscience, catalog number: 65-0865-14 )
- Low endotoxin fetal bovine serum (FBS, heat-inactivated for 20 min at 56 °C) (Biowest, catalog number: S1860 )
- Low endotoxin BSA (fraction V) (Euromedex, catalog number: UA1315 )
- CFSE (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: C34554 )
- Liberase TM (Roche Diagnostics, catalog number: 05401119001 )
- DNase I (Roche Diagnostic, catalog number: 04536282001 )
- Red blood cell lysis buffer (Sigma-Aldrich, catalog number: R7757 )
- EasySepTM Mouse Pan-DC Enrichment Kit (STEMCELL Technologies, catalog number: 19763 )
- EasySepTM Mouse Naïve CD8+ T Cell Isolation Kit (STEMCELL Technologies, catalog number: 19858 )
- FACS antibodies (all anti-mouse):
- CD69-eFluor® 450 (clone H1.2F3, dilution 1/300) (Affymetrix, eBioscience, catalog number: 48-0691-82 )
- CD8a-PerCP-Cy5.5 (clone 53-6.7, dilution 1/300) (Affymetrix, eBioscience, catalog number: 45-0081-82 )
- TCR vβ 5.1-PE (clone MR9-4, dilution 1/500) (BD, PharmingenTM, catalog number: 553190 )
- CD25-FITC (clone 7D4, dilution 1/200) (BD, PharmingenTM, catalog number: 553072 )
- CD4-PE-Cy7 (clone RM4-5, dilution 1/300) (BD, PharmingenTM, catalog number: 552775 )
- CD25-APC (clone PC61.5, dilution 1/300) (Affymetrix, eBioscience, catalog number: 17-0251-81 )
- CD19 eFluor® 450 (clone 1D3, dilution 1/500) (Affymetrix, eBioscience, catalog number: 48-0193 )
- CD3 eFluor® 450 (clone 17A2, dilution 1/500) (Affymetrix, eBioscience, catalog number: 48-0032-80 )
- CD11c-FITC (clone HL3, dilution 1/500) (BD, PharmingenTM, catalog number: 553801 )
- CD11c-APC (clone N418, dilution 1/400) (Affymetrix, eBioscience, catalog number: 17-0114 )
- CD8-PE (clone 53-6.7, dilution 1/500) (BD, PharmingenTM, catalog number: 553032 )
- CD11b-PE (clone M1/70, dilution 1/300) (Affymetrix, eBioscience, catalog number: 12-0112 )
- CD40-PE (clone 3/23, dilution 1/150) (BD, PharmingenTM, catalog number: 553791 )
- CD86-PE (clone GL1, dilution 1/300) (Affymetrix, eBioscience, catalog number: 12-0862 )
- MHC Class II I-Ab-PE (clone AF6-120.1, dilution 1/300) (Affymetrix, eBioscience, catalog number: 12-5320 )
- CD69-eFluor® 450 (clone H1.2F3, dilution 1/300) (Affymetrix, eBioscience, catalog number: 48-0691-82 )
- BMDC culture medium (see Recipes)
- B3Z and T cell culture (see Recipes)
- Digestion medium for spleens (see Recipes)
- MACS buffer (see Recipes)
Equipment
- Incubator (37 °C and 5% CO2)
- Refrigerated centrifuge for tubes of 2 ml, 15 ml and 50 ml size as well as 96-well plates
- Stuart test tube rotator wheel tolerating 4 °C (Bibby Scientific, model: SB3 )
- Multicolor flow cytometer (MAQSquant, Miltenyi; FACSverse, Becton Dickinson or similar)
- Multicolor FACS sorter (for example, BD, model: FACSAria III )
- Fluorometric plate reader (OD: 590 nm)
Software
- FlowJo 10 software (FlowJo, LLC.)
- Prism 6 software (GraphPad Software, Inc.)
Procedure
Part I. Cross-presentation analysis of bead-bound OVA in BMDC in vitro
- Preparation of beads (Day: -1, sterile environment)
The day before the cross-presentation experiment, particles are coupled to OVA (bead-bound OVA: bbOVA). Polystyrene beads of 3 μm diameter are coated with soluble OVA and soluble BSA ranging from 100% OVA (most concentrated) to 100% BSA (background). Since BSA is not cross-presented, the preparation of beads coated with 100% BSA is the accurate control to check whether beads without any antigen coating are able to induce any additional activation of DC.- Estimate the amount of 3 μm beads that will be used during the cross-presentation experiment on day 0. Consider to prepare a minimum of at least four different conditions:
- 100% OVA and 0% BSA: 10 mg/ml OVA
- 50% OVA and 50% BSA: 5 mg/ml OVA + 5 mg/ml BSA
- 25% OVA and 75% BSA: 2.5 mg/ml OVA + 7.5 mg/ml BSA
- 0% OVA and 100% BSA: 10 mg/ml BSA
- Dilute the beads differentially depending on the readout. For B3Z T cell hybridomas, beads are diluted 1/10 (which corresponds to a bead/cell ratio of approximately 200), while for OT-I T cells the dilution is 1/25 (which corresponds to a bead/cell ratio of 600).
The beads are added to BMDC in a final volume of 100 μl. In order to analyze cross-presentation in 10 samples with the 3 different read-outs of this protocol, you will need per condition:
10 samples for B3Z → 1 ml of culture medium → 100 μl of beads (1/10 dilution)
10 samples for OT-I CD69 expression → 40 μl of beads (1/25 dilution)
10 samples for OT-I proliferation → 40 μl of beads (1/25 dilution)
Therefore, a final bead volume of 200-250 μl is needed for 10 samples. - Put a maximum of 250 μl of beads per 2 ml Eppendorf tube, and wash them with 1.5 ml of PBS. Centrifuge at 19,000 x g for 3 min at 4 °C. Repeat washing twice.
Note: If you are assessing 10 samples with the 3 readouts, and doing 4 different conditions of bbOVA (100%, 50%, 25% and 0%) you should have at this point 4 tubes with 200-250 μl of beads (1 for each condition). - Discard supernatant, and resuspend the beads in the different protein suspensions as shown in the following (250 μl of beads per 750 μl of soluble protein mix):
Stock concentration of OVA: 50 mg/ml (Working concentration: 10 mg/ml)
Stock concentration of BSA: 50 mg/ml (Working concentration: 10 mg/ml)
Cond. 1) 100% OVA: 0% BSA → 150 μl of OVAstock + 600 μl PBS
Cond. 2) 50% OVA: 50% BSA → 75 μl of OVAstock + 75 μl of BSAstock + 600 μl PBS
Cond. 3) 25% OVA: 75% BSA → 37.5 μl of OVAstock + 112.5 μl of BSAstock + 600 μl PBS
Cond. 4) 0% OVA: 100% BSA → 150 μl of BSAstock + 600 μl PBS - Incubate overnight at 4 °C with permanent agitation on a rotator wheel.
- Washing the beads (Day: 0, sterile environment)
- Spin the bbOVA with 19,000 x g for 5 min at 4 °C. Subsequently, discard supernatant with a pipette trying not to touch the beads.
- Resuspend the bbOVA in 1 ml of cold PBS by pipetting. Centrifuge at 19,000 x g for 5 min at 4 °C.
- Repeat washing step twice.
- Resuspend the bbOVA in the original volume (200-250 μl in this example). The beads are ready to use. Keep bbOVA on ice.
- Spin the bbOVA with 19,000 x g for 5 min at 4 °C. Subsequently, discard supernatant with a pipette trying not to touch the beads.
- Loading BMDC with bbOVA, Co-culture with T cells (Day: 0, sterile environment)
Plate in 96-well round bottom plates: 100,000 BMDC per well for B3Z readout and 10,000 BMDC per well for both OT-I readouts (Figure 1).
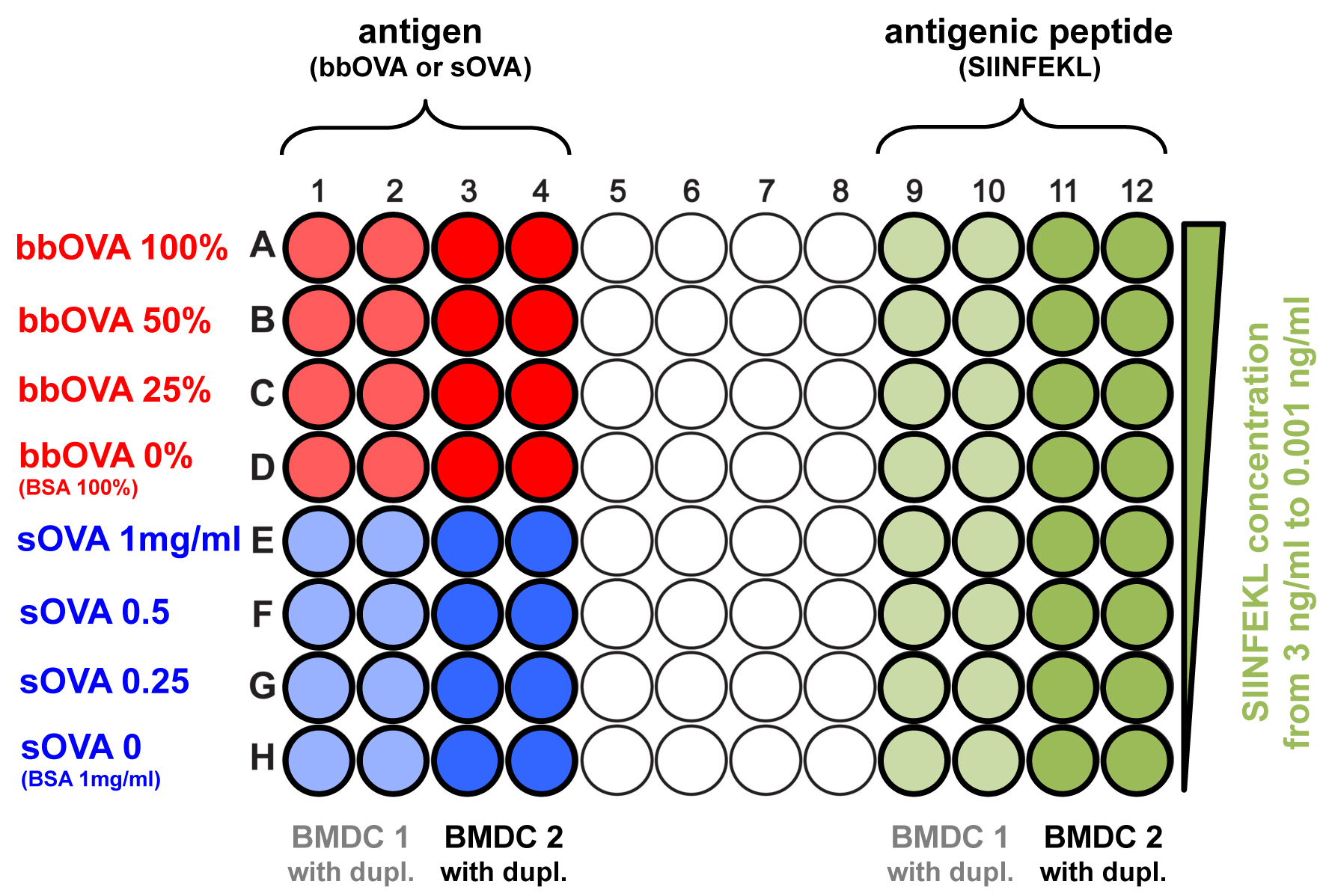
Figure 1. Scheme of an average 96-well plate loaded with BMDCs. Plate 100,000 BMDCs/well in 50 μl of BMDC culture medium for B3Z readout, or 10,000 cells/well in 50 μl of BMDC culture medium for OT-I readouts. Add the antigen to the right concentration in 50 μl of BMDC culture medium for a final volume of 100 μl. In the scheme, sOVA and SIINFEKL peptide are also analyzed (see Notes 2 and 3). In the scheme, a minimum setup is shown: two different BMDC samples are analyzed, which are plated in duplicates. Additional replicates are suggested to increase robustness of the obtained data.- The correct amount of BMDC is plated in 50 μl of BMDC culture medium per well.
- The final dilution of bbOVA in BMDC culture medium is, as mentioned before: 1/10 for B3Z and 1/25 for OT-I. Please consider that 50 μl of bbOVA in BMDC culture medium will be added to 50 μl of BMDC for a final volume of 100 μl per well. Thereby, bbOVA concentration must be twice the final dilution (1/5 for B3Z, 1/12.5 for OT-I).
- For the OT-I T cell proliferation readout, incubate BMDC with bbOVA for 1 h at 37 °C. Consequently, cross-presenting BMDC are co-cultured with 60,000-100,000 CFSE-stained OT-I T cells and T cell proliferation is analyzed 72 h later (continues in step 5a).
- For B3Z and OT-I CD69 expression readouts, incubate BMDC with bbOVA for 4-5 h at 37 °C.
- Wash the cross-presenting BMDC with 100 μl/well of 0.1% PBS-BSA (always keep reagents and solutions on ice) and centrifuge at 800 x g for 2 min at 4 °C. Flick the plate and repeat washing twice.
- Fix cross-presenting BMDC: add 50 μl/well of freshly made PBS-glutaraldehyde (GTA) 0.008% (vol/vol) and incubate for 5 min on ice. Mix cells and PBS-GTA by pipetting.
- Add 50 μl of PBS-glycine 0.4 M to the PBS-GTA 0.008% solution, and centrifuge at 800 x g for 2 min at 4 °C. Flick the plate.
- Add 100 μl of PBS-glycine 0.2 M and centrifuge at 800 x g for 2 min at 4 °C. Flick the plate.
- Wash twice with 200 μl/well of B3Z and T cell culture medium (see Recipes). Centrifuge again and flick the plate.
- Resuspend fixed, cross-presenting BMDC in 100 μl/well of cell culture medium.
- Add 60,000-100,000 effector cells (B3Z or OT-I, see Notes 4 and 5) per well in 100 μl cell culture medium (to a final volume of 200 μl).
- Incubate for 16 h at 37 °C for B3Z T cells or for 18-20 h at 37 °C for OT-I T cells.
- The correct amount of BMDC is plated in 50 μl of BMDC culture medium per well.
- Analysis of B3Z read-out and CD69 expression of OT-I cells (Day: 1)
- B3Z measurement:
- Centrifuge at 800 x g for 2 min at 4 °C. Supernatant can be kept for subsequent cytokine analysis.
- Wash once with 100 μl/well of 0.1% PBS-BSA, spin down and flick.
- Add 120 μl/well of CPRG (Chlorophenol red-β-D-galactopyranoside). Mix properly to resuspend the B3Z cells.
- Incubate at 37 °C and check every 30 min until the solution changes its color to orange/red (usually, after 2 h).
- Measure OD at 590 nm every 30-60 min until the reaction gets saturated (after 4-6 h).
- Measurement of CD69 expression of OT-I cells:
- Centrifuge at 800 x g for 2 min at 4 °C. Supernatant can be kept for subsequent cytokine analysis.
- Wash once with 100 μl/well of 0.1% PBS-BSA, spin down and flick the plate.
- Stain the cells for 15 min at 4 °C with 100 μl/well of fixable viability dye eFluor 780 + Fc block (1/100) in PBS. Centrifuge and flick the plate.
- Wash once with 0.1% PBS-BSA, centrifuge and flick the plate.
- Stain the cells for 40 min at 4 °C with 70 μl/well of a combination of CD69-eFluor450, CD8a-PerCP-Cy5.5, TCR vβ 5.1-PE, CD25-FITC and CD4-PE-Cy7 diluted in 1% PBS-BSA (dilution factors are provided in Materials and Reagents). Centrifuge and flick the plate.
- Wash 3 times with 0.1% PBS-BSA, centrifuge and flick the plate.
- Resuspend in 100 μl/well of 0.1% PBS-BSA and analyze by flow cytometry (see Figure 2a).
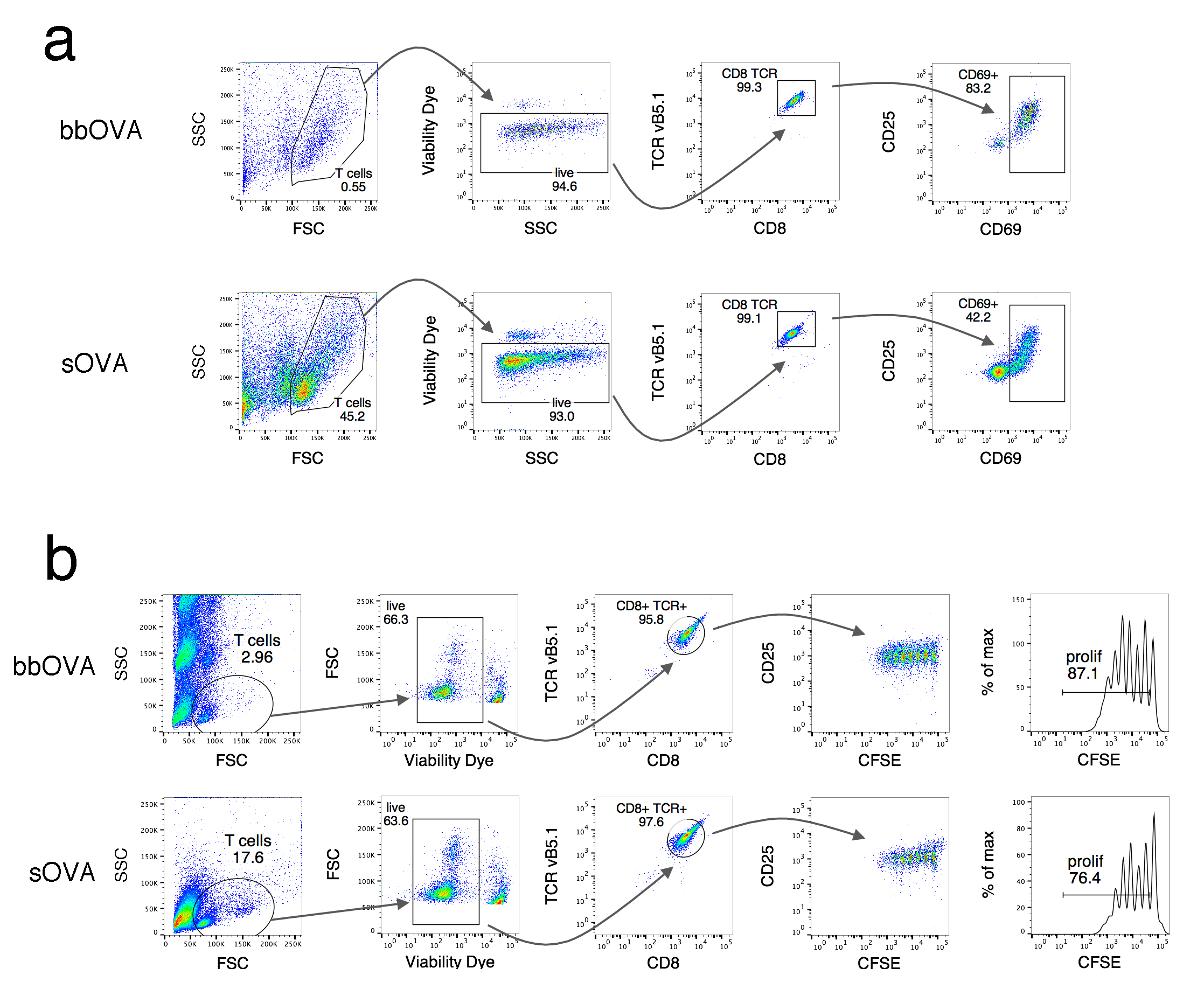
Figure 2. Gating strategy for cross-presentation analysis. a. Activation of OT-I T cells by measuring CD69 expression at the cell surface. BMDC were incubated 5 h with bbOVA (upper panel) and sOVA (see Note 2) (lower panel), fixed and co-cultured with OT-I T cells. After 16 h, CD69 expression was measured. b. Proliferation of OT-I T cells. BMDC were incubated for 1 h in the presence of bbOVA (upper panel) and sOVA (lower panel) and co-cultured for 72 h with CFSE-labeled OT-I T cells. Proliferation was addressed by measuring loss of CFSE staining by flow cytometry and by calculating the proliferation index (right panel).
- Analysis of OT-I T cell proliferation (Day: 3)
- Centrifuge at 800 x g for 2 min at 4 °C. Supernatant can be kept for subsequent cytokine analysis.
- Wash once with 100 μl/well of 0.1% PBS-BSA, spin down and flick the plate.
- Stain the cells for 15 min at 4 °C with 100 μl/well of fixable viability dye eFluor 780 + Fc block (1/100) in PBS. Centrifuge and flick the plate.
- Wash once with 0.1% PBS-BSA, centrifuge and flick the plate.
- Stain the cells for 40 min at 4 °C with 70 μl/well of a combination of CD8a-PerCP-Cy5.5, TCR vβ 5.1-PE, CD25-APC and CD4-PE-Cy7 diluted in 1% PBS-BSA (dilution factors are provided in Materials and Reagents). Centrifuge and flick the plate.
- Wash 3 times with 0.1% PBS-BSA, centrifuge and flick the plate.
- Resuspend in 100 μl/well of 0.1% PBS-BSA and analyze by flow cytometry (see Figure 2b).
- Centrifuge at 800 x g for 2 min at 4 °C. Supernatant can be kept for subsequent cytokine analysis.
Part II. Cross-presentation of bbOVA by splenic CD8+ dendritic cells, analyzed ex vivo
In this protocol, blue dyed beads coated with OVA are injected intravenously into mice. Subsequently, after DC have internalized the particulate antigen and cross-presented it in vivo, spleens are harvested and digested. CD11c+ cells from whole digested spleens are then purified, and the enriched CD11c+ fraction is stained. CD8+ DC that have phagocytosed beads are sorted and co-cultured with OT-I T cells for further analysis (Figure 3).
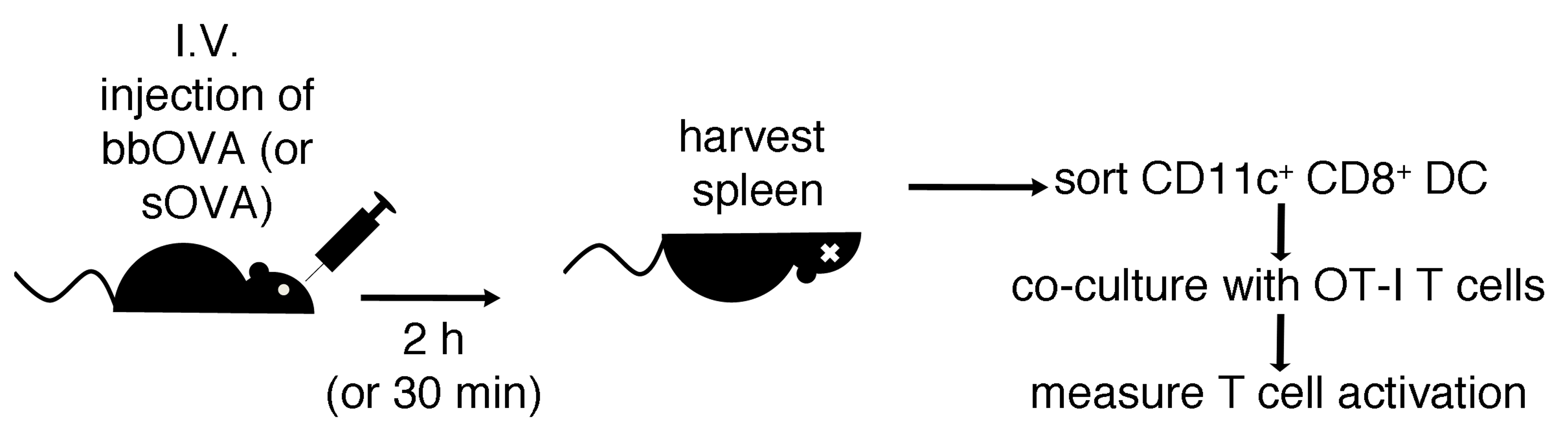
Figure 3. Workflow of the antigen cross-presentation approach in vivo
- Preparation of beads (Day: -1)
- Calculate the number of blue dyed 1.0 μm beads that will be used. 4.5 x 109 beads are injected per mouse (bead stock solution is around 4.5 x 1010 particles/ml). Therefore, 100 μl per mouse are injected.
- Wash the beads with 1.5 ml cold PBS (again 250 μl of beads per 2 ml Eppendorf tube). Centrifuge at 19,000 x g for 5 min at 4 °C. Repeat.
- Resuspend each 250 μl of beads in 500 μl of OVA 50 mg/ml, and pool them (if needed) into a 15 ml centrifuge tube. Incubate overnight at 4 °C with permanent agitation on a rotator wheel.
- Calculate the number of blue dyed 1.0 μm beads that will be used. 4.5 x 109 beads are injected per mouse (bead stock solution is around 4.5 x 1010 particles/ml). Therefore, 100 μl per mouse are injected.
- Washing the beads (Day: 0)
- For washing purposes, divide the beads in 2 ml Eppendorf tubes (250 μl per tube).
- Spin the bbOVA at 19,000 x g at 4 °C for 5 min. Subsequently, discard supernatant with a pipette, trying not to touch the beads.
- Resuspend the bbOVA in 1.5 ml of cold PBS. Centrifuge at 19,000 x g at 4 °C for 5 min.
- Repeat washing step twice.
- Resuspend the bbOVA in the original volume (250 μl in this example) and pool them. The beads are ready to use. Store them on ice.
- For washing purposes, divide the beads in 2 ml Eppendorf tubes (250 μl per tube).
- Injection of mice, purification of DC and co-culture with T cells (Day: 0) (see Figure 3)
- Inject 100 μl of beads per mouse intravenously (either in the eye vein or in the tail). Use Terumo 1 ml insulin syringes with 25 G needle. Wait 2 h (time for the bbOVA to be phagocytosed and cross-presented).
- Sacrifice the mice and harvest spleens.
- Flush the spleens in non-treated 6-well plates with 2 ml of digestion medium (see Recipes) per spleen by using 25 G needles and 2.5 ml syringes. Incubate for 15 min at 37 °C.
- Mince the spleens with scalpel blades in the same well and incubate for another 15 min. Stop Liberase/DNAse reaction by adding 2 ml of FBS.
- Smash and filter the digested spleens through the cell strainer with the flat part of a syringe plunger into 50 ml centrifuge tubes. Centrifuge and discard supernatant.
- Lyse red blood cells at RT (5 min of 2 ml of red blood cell lysis buffer per spleen is sufficient). Add 30 ml of PBS and centrifuge (350 x g, 5 min, 4 °C).
- Discard supernatant and resuspend each spleen in 600 μl of cold sorting medium (PBS + 0.5 % FBS). Pass the cells through the suggested Miltenyi pre-separation filters (or similar) and collect them in 14 ml tubes for magnetic purification.
- In order to isolate CD11c+ cells, we use the EasySep Mouse Pan-DC Enrichment Kit, but any other negative selection-based enrichment procedure might be used. Follow the manufacturer’s indications.
- Stain the CD11c+-enriched cell fraction for 40 min at 4 °C with 70 μl/well of a combination of CD19-eFluor 450, DAPI, CD3-eFluor 450, CD11c-FITC and CD8-PE (see Note 2).
- Sort the cells following the gating strategy shown in Figure 4.
- Plate phagocytic CD8+ DC at different amounts to obtain three different, final DC/T cell ratios:
30,000 DC/100,000 T cells
3,000 DC/100,000 T cells
300 DC/100,000 T cells - Fix CD8+ DC with 0.008% PBS-GTA as described before (Procedure of part I).
- Co-culture fixed DCs with 100,000 OT-I T cells to a final volume of 200 μl of B3Z and T cell culture medium.
- For T cell analysis, proceed as described before (T cell read-outs from Procedure of part I).
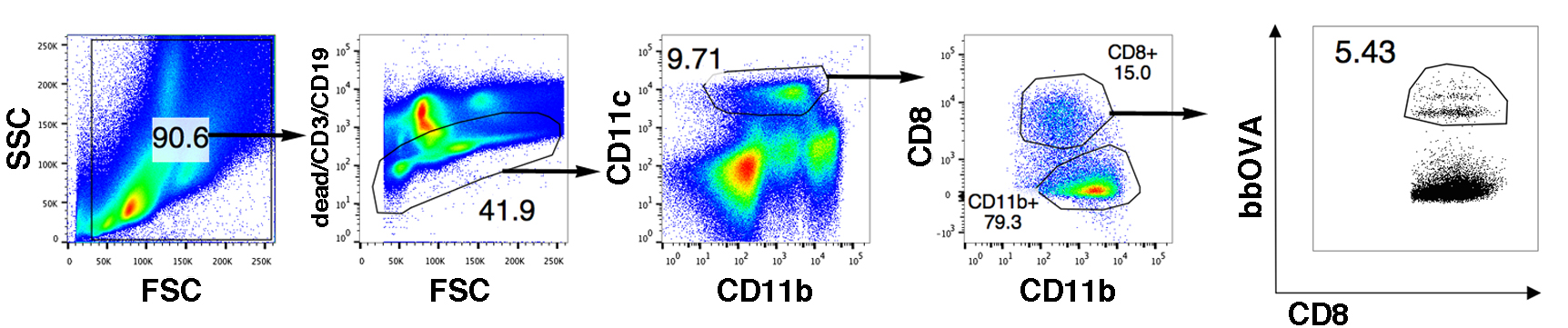
Figure 4. Gating strategy to sort CD8+ DC from spleen. Total splenic cells were gated in order to exclude B, T and dead cells. Subsequently, gates were performed on CD11c+ and CD8+ cells. CD11c+ CD8+ cells that have phagocytosed at least one bead were sorted and co-cultured with OT-I T cells.
- Inject 100 μl of beads per mouse intravenously (either in the eye vein or in the tail). Use Terumo 1 ml insulin syringes with 25 G needle. Wait 2 h (time for the bbOVA to be phagocytosed and cross-presented).
Data analysis
Each single experiment needs to be performed with a sufficient number of technical replicates (at least in triplicates). Conclusions can be drawn from the analysis of an appropriate number of biological replicates. The results shown here were confirmed by a minimum of 3 independent experiments. Analysis of flow cytometry data was performed using FlowJo 10 software (FlowJo, LLC.). Statistical analysis was performed using Prism 6 software (GraphPad Software, Inc.).
Notes
Part I. Cross-presentation analysis of bead-bound OVA in BMDC in vitro
- BMDC are generated by culture in GM-CSF-containing medium (see Recipes) for 10 days. Percentages of CD11c and CD11b are always higher than 85% in used cultures.
- Cross-presentation of soluble OVA (sOVA) can be measured in parallel. In our lab, we generally use sOVA ranging from 2 mg/ml to 0 mg/ml (4 different concentrations are usually sufficient).
- The best control for cross-presentation, to our knowledge, is the OVA peptide 257-264 (SIINFEKL). This peptide is incorporated into the cell and loaded onto MHC class I molecules without being processed. We strongly recommend using such a control in different dilutions (a range from 3 ng/ml to 0.001 ng/ml is sufficient) for all cross-presentation assays.
- Culturing B3Z T cells: Grow B3Z T cells in B3Z and T cell culture medium (see Recipes), split cells every 2-3 days; detach the cells manually by hitting smoothly against the flask. The day before the cross-presentation experiment, seed them at a concentration of 0.1 x 106 cells/ml. Co-culture them with cross-presenting BMDC. After 16 h, T cell activation is measured detecting β-galactosidase activity by optical density at 590 nm using CPRG as substrate for the reaction.
- In order to get OT-I T cells for co-culture with BMDC, we use kits for mouse naïve CD8+ T cell purification (obtained either from Stemcell or Miltenyi) of spleens and lymph nodes of OT-I mice. We use MACS buffer (recipes 4) for washing purposes.
- To obtain CFSE-labeled OT-I T cells, resuspend 1 x 107 cells/ml of CD8+ T cells and label them with 10 μM of CFSE proliferative dye for 10 min at room temperature in the dark. Wash cells once with 10 ml of PBS and resuspend in sterile PBS to a concentration of 1 x 107 cells/ml.
Part II. Cross-presentation of bbOVA by splenic CD8+ dendritic cells, analyzed ex vivo
- Cross-presentation of sOVA can be measured in parallel. Consider injecting 100 μl of OVA (i.v.) at a concentration of 20 mg/ml. Wait for 30 min instead of 2 h and harvest spleens to purify cross-presenting DC.
- With the labeling of CD19, CD3 and DAPI, it is possible to exclude B, T and dead cells, respectively, from the analysis. Be aware that blue dyed beads are visible in both, the APC and the APC-Cy7 channel. No fluorophore should be used in those channels.
- Consider that the proportion of phagocytic CD8+ DC in spleen is really low. You should get around 100,000-200,000 cells per spleen. While this amount is enough for OT-I T cells co-culture (to analyze both OT-I CD69 expression and CFSE OT-I proliferation), co-culture with B3Z T cells might require the sacrifice of many more mice as well as many hours of cell sorting. By the time you will get sufficient cells for B3Z analysis, they might be already dead. Therefore, we suggest to not address this readout for ex vivo analysis of cross-presentation.
Recipes
- BMDC culture medium
IMDM containing 10% heat-inactivated FBS
100 IU/ml penicillin
100 μg/ml streptomycin
2 mM glutamax
50 μM β-mercaptoethanol
1x MEM non-essential amino acids
1x sodium pyruvate
Supernatant from J558 plasmacytoma cells was used as GM-CSF source (Winzler et al., 1997). - B3Z and T cell culture
RPMI containing 10% heat-inactivated FBS
100 IU/ml penicillin
100 μg/ml streptomycin
2 mM glutamax
50 μM β-mercaptoethanol
1x MEM non-essential amino acids
1x sodium pyruvate - Digestion medium for spleens
Liberase 0.2 mg/ml (resuspend one bottle of 5 mg Liberase in 3 ml of RPMI-1640, which is sufficient for 15 spleens; use 0.2 ml of this solution per spleen) and DNase I 0.1 mg/ml in RPMI-1640 (final volume: 1.5 ml per spleen). - MACS buffer
1x PBS
2% FBS (or 2% BSA)
5 mM EDTA
Acknowledgments
This work was supported by the French National Research Agency through the ‘Investments for the Future’ program (France-BioImaging, ANR-10-INSB-04), ANR-11-LABX-0043 and by the CelTisPhyBio Labex (N- ANR-10-LBX-0038), part of the IDEX PSL (ANR-10-IDEX-0001-02 PSL). We are grateful to the financial support by the European Research Council (2013-AdG No.340046 DCBIOX), by La Ligue Nationale contre le Cancer (EL2014.LNCC/SA), by Fonds Wetenschappelijk Onderzoek (FWO; 1526615N), by an EMBO long-term fellowship (ALTF 883-2011) and by fellowships of Fondation Recherche Médicale (SPF20101221176) and the omics@VIB program (co-financed by the Marie Curie FP7 People Cofund).
References
- Alloatti, A., Kotsias, F., Pauwels, A. M., Carpier, J. M., Jouve, M., Timmerman, E., Pace, L., Vargas, P., Maurin, M., Gehrmann, U., Joannas, L., Vivar, O. I., Lennon-Dumenil, A. M., Savina, A., Gevaert, K., Beyaert, R., Hoffmann, E. and Amigorena, S. (2015). Toll-like receptor 4 engagement on dendritic cells restrains phago-lysosome fusion and promotes cross-presentation of antigens. Immunity 43(6): 1087-1100.
- Crozat, K., Tamoutounour, S., Vu Manh, T. P., Fossum, E., Luche, H., Ardouin, L., Guilliams, M., Azukizawa, H., Bogen, B., Malissen, B., Henri, S. and Dalod, M. (2011). Cutting edge: expression of XCR1 defines mouse lymphoid-tissue resident and migratory dendritic cells of the CD8α+ type. J Immunol 187(9): 4411-4415.
- Dorner, B. G., Dorner, M. B., Zhou, X., Opitz, C., Mora, A., Guttler, S., Hutloff, A., Mages, H. W., Ranke, K., Schaefer, M., Jack, R. S., Henn, V. and Kroczek, R. A. (2009). Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity 31(5): 823-833.
- Joffre, O. P., Segura, E., Savina, A. and Amigorena, S. (2012). Cross-presentation by dendritic cells. Nat Rev Immunol 12(8): 557-569.
- Karttunen, J., Sanderson, S. and Shastri, N. (1992). Detection of rare antigen-presenting cells by the lacZ T-cell activation assay suggests an expression cloning strategy for T-cell antigens. Proc Natl Acad Sci U S A 89(13): 6020-6024.
- Kurts, C., Heath, W. R., Carbone, F. R., Allison, J., Miller, J. F. and Kosaka, H. (1996). Constitutive class I-restricted exogenous presentation of self antigens in vivo. J Exp Med 184(3): 923-930.
- Winzler, C., Rovere, P., Rescigno, M., Granucci, F., Penna, G., Adorini, L., Zimmermann, V. S., Davoust, J. and Ricciardi-Castagnoli, P. (1997). Maturation stages of mouse dendritic cells in growth factor-dependent long-term cultures. J Exp Med 185(2): 317-328.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Alloatti, A., Kotsias, F., Hoffmann, E. and Amigorena, S. (2016). Evaluation of Cross-presentation in Bone Marrow-derived Dendritic Cells in vitro and Splenic Dendritic Cells ex vivo Using Antigen-coated Beads. Bio-protocol 6(22): e2015. DOI: 10.21769/BioProtoc.2015.
Category
Immunology > Immune cell function > Dendritic cell
Cell Biology > Cell-based analysis
Cell Biology > Cell-based analysis > Flow cytometry
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











