- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Hydrogen Peroxide Measurement in Arabidopsis Root Tissue Using Amplex Red
Published: Vol 6, Iss 21, Nov 5, 2016 DOI: 10.21769/BioProtoc.1999 Views: 16362
Reviewed by: Arsalan DaudiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Heterologous Production of Artemisinin in Physcomitrium patens by Direct in vivo Assembly of Multiple DNA Fragments
Nur Kusaira Khairul Ikram [...] Henrik Toft Simonsen
Jul 20, 2023 2333 Views

Utilizing FRET-based Biosensors to Measure Cellular Phosphate Levels in Mycorrhizal Roots of Brachypodium distachyon
Shiqi Zhang [...] Maria J. Harrison
Jan 20, 2025 2389 Views

High-Performance Liquid Chromatography Quantification of Glyphosate, Aminomethylphosphonic Acid, and Ascorbate in Culture Medium and Microalgal Cells
Juan Manuel Ostera [...] Gabriela Malanga
Apr 5, 2025 1182 Views
Abstract
This protocol describes the measurement of hydrogen peroxide (H2O2) content in Arabidopsis root tissue by using the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit. When root tissue is disrupted and resuspended in phosphate buffer, H2O2 is released from the cells. The obtained root extracts containing H2O2 can be mixed with a solution containing Amplex® Red reagent (10-acetyl-3,7-dihydrophenoxazine). In the presence of horseradish peroxidase, the Amplex® Red reagent reacts with H2O2 in a 1:1 stoichiometry. The resulting product is the red-fluorescent compound resorufin which can be detected fluorometrically or spectrophotometrically. Our protocol is based on the manual of the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit and describes a step-by-step procedure with a detailed description of the necessary controls and data analysis. We have also included modifications of the protocol, notes and examples that intend to aid the user in easily reproducing the assay with their own samples.
Background
Reactive oxygen species (ROS), such as H2O2, can be generated in the cell as a result of a developmental cue or a stress condition. In high amounts, ROS accumulation can be detrimental by causing cellular damage. However, increased ROS production can also have a signaling role and serve as a secondary messenger in controlling downstream cellular responses. In plants, a signaling role for ROS has been shown for many abiotic stresses, such as drought, salinity, temperature stress, and nutrient deprivation (Mittler, 2002; Mittler and Blumwald, 2015; Xia et al., 2015). In our recent publication Le et al. (2016), we have investigated the connection between ROS production and iron (Fe) deficiency response regulation by investigating the H2O2 content of roots from wild type and mutant Arabidopsis plant lines grown under sufficient and deficient Fe supply conditions.
Materials and Reagents
- Kimtech® science precision tissues (Carl Roth, catalog number: AA63.1 )
- 2 ml microcentrifuge tubes, safe-seal (SARSTEDT, catalog number: 72.695.500 )
- Pipet tips
- Aluminium foil
- 96-well microtiter plates for absorbance measurement (e.g., UV-Star® microplate, 96 well, half area, µClear® [Greiner Bio-One, catalog number: 675801 ])
- 96-well microtiter plates for fluorescence measurement (e.g., 96 well, half area, black [Greiner Bio-One, catalog number: 675076 ])
- Arabidopsis seeds
- Sterile distilled water
- Plant agar (Duchefa Biochemie, catalog number: P1001.1000 )
- Liquid nitrogen
- Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit (Thermo Fisher Scientific, Molecular ProbesTM, catalog number: A22188 )
- H2O2 working solution
- HRP enzyme
- Horseradish peroxidase
- Sodium hypochlorite (NaOCl) (Carl Roth, catalog number: 9062.3 )
- Triton X-100 (SERVA Electrophoresis, catalog number: 37238 or Sigma-Aldrich, catalog number: X-100 )
- Magnesium sulfate heptahydrate (MgSO4·7H2O) (Sigma-Aldrich, catalog number: M9397 )
Note: This product has been discontinued. Alternatively, MgSO4·7H2O from Carl Roth can be used (Carl Roth, catalog number: T888 ) - Potassium dihydrogen phosphate (KH2PO4) (Sigma-Aldrich, catalog number: P0662 ; or Carl Roth, catalog number: 3904 )
- Potassium nitrate (KNO3) (Carl Roth, catalog number: P021.2 )
- Calcium nitrate tetrahydrate [Ca(NO3)2·4H2O] (Carl Roth, catalog number: P740.2 )
- Potassium chloride (KCl) (Carl Roth, catalog number: 6781.1 )
- Boric acid (H3BO3) (Carl Roth, catalog number: 6943.1 )
- Manganese sulfate (MnSO4) (AppliChem, catalog number: A1038 ; or Carl Roth, catalog number: X890.1 )
- Zinc sulfate heptahydrate (ZnSO4·7H2O) (VWR, catalog number: VWRC29253.236 )
- Copper sulfate pentahydrate (CuSO4·5H2O) (Thermo Fisher Scientific, Fisher Scientific, catalog number: AC197720050 )
- Ammonium heptamolybdate tetrahydrate [(NH4)6Mo7O24·4H2O] (Sigma-Aldrich, catalog number: 431346 ; or Carl Roth, catalog number: 7311 )
- D(+)-sucrose (Carl Roth, catalog number: 4621.1 )
- Ferric sodium ethylenediaminetetraacetic acid (FeNaEDTA) (Carl Roth, catalog number: 8043.1 )
- 3-(2-Pyridyl)-5,6-diphenyl-1,2,4-triazine-4’,4’’-disulfonic acid sodium salt (Ferrozine) (Sigma-Aldrich, catalog number: 82950 )
- Sterilization solution (see Recipes)
- Hoagland medium (see Recipes)
- Phosphate buffer (see Recipes)
Equipment
- Tube rotator (e.g., VWR, catalog number: 10136-084 )
- Mortars and pestles (e.g., MTC Haldenwanger)
- Centrifuge
- Multi-channel pipet
- Analytical scales, e.g., ALJ160_4NM (Kern)
- Benchtop centrifuge with cooling (e.g., Thermo Fisher Scientific, Thermo ScientificTM, model: Heraeus Fresco 21 Centrifuge )
- Plate reader (e.g., Tecan Trading, model: Infinite® M200 Pro )
Note: The plate reader should be able to excite in the range of 530-560 nm and detect fluorescence at approx. 590 nm, or detect absorbance at approx. 560 nm. - Homogenizer (e.g., such as Precellys® 24 homogenizer) (VWR, catalog number: 432-3750 )
Software
- Microsoft Excel
Procedure
A graphical overview of the different protocol steps starting with sample preparation and finishing with resorufin detection is provided in Figure 1.
Figure 1. A graphical overview of the different protocol steps for H2O2 measurement describing the preparation of samples, H2O2 standards and negative controls, which are color coded depending on their identity. The same color code is used for the plate setup in Figure 3.
- Plant extract preparation for H2O2 content measurement
- Grow Arabidopsis plants to the desired age under the desired growth conditions.
- In our case, Arabidopsis seeds were surface sterilized as described previously (Lingam et al., 2011). The seeds were incubated for 8 min in sterilization solution (see Recipes) at room temperature on a tube rotator and subsequently washed five times with sterile distilled water. After the fifth wash, the seeds were stored in 0.1% plant agar for 1-2 days in the dark at 4 °C for stratification.
Note: Do not incubate the seeds in the sterilization solution for longer than 8 min since this can lead to a reduced seed viability. - Seeds were plated out and germinated on upright Hoagland medium agar plates (see Recipes). Seedlings were grown under long-day conditions (16 h light/21 °C and 8 h dark/19 °C) for 5, 7, 8 and 10 days before harvesting (an example of plates with 10-day-old seedlings is shown in Figure 2). Roots were separated from the shoot directly on the agar plate on which the plants were growing (in order to avoid drying of the roots) and quickly collected into a bunch. The bunch of roots was then very briefly (for a second) and gently laid on a Kimtech precision paper to remove excess moisture before freezing it in liquid nitrogen.

Figure 2. Example of wild-type Arabidopsis plants (Col-0) grown on upright Hoagland medium agar plates for 10 days. The plants were either grown continuously on sufficient (left) or deficient (right) Fe supply. Scale bars = 1 cm.
- In our case, Arabidopsis seeds were surface sterilized as described previously (Lingam et al., 2011). The seeds were incubated for 8 min in sterilization solution (see Recipes) at room temperature on a tube rotator and subsequently washed five times with sterile distilled water. After the fifth wash, the seeds were stored in 0.1% plant agar for 1-2 days in the dark at 4 °C for stratification.
- Grind frozen roots with mortar and pestle in liquid nitrogen.
- Collect the frozen tissue powder in pre-cooled 2 ml microcentrifuge tubes and weigh the samples on analytical scales.
- Add 200 µl of ice-cold phosphate buffer (see Recipes) to 30 mg of tissue powder and resuspend on ice. If the amount of powder differs adjust the buffer volume accordingly. For example, for 15 mg powder add 100 µl of phosphate buffer. In order to avoid plant material loss due to sticking to pipet tips we do not resuspend the powder by pipetting up and down. Rather, we let the samples on ice, with the added buffer, for not more than a minute under the powder is well soaked. Afterwards, we flip the microcentrifuge tubes until the suspension appears homogenous.
Note: The minimal amount of root powder that we have been able to use successfully has been 15 mg, which in our growth conditions corresponded to approx. 25 10-day-old seedlings. - Centrifuge for 3 min at 16,200 x g (corresponds to 13,000 rpm in the centrifuge suggested in Equipment), 4 °C. Transfer the supernatant to a new pre-cooled microcentrifuge tube. This is the sample that will be used for the H2O2 assay (see Figure 3A for an example of sample identity). Keep the samples on ice and proceed, at best, immediately with the H2O2 measurement. We do not have data on how stable the H2O2 in the obtained extracts is.
- Grow Arabidopsis plants to the desired age under the desired growth conditions.
- H2O2 measurement
- Prepare 10 mM Amplex® Red reagent stock solution, 1x reaction buffer (0.05 M sodium phosphate buffer, pH 7.4), 10 U/ml horseradish peroxidase (HRP) stock solution, and 20 mM H2O2 working solution according to the instructions provided with the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit.
Notes:- The Amplex® Red reagent is air and light sensitive. The 1x reaction buffer is stable at room temperature. The HRP stock solution should be aliquoted and kept at -20 °C. The 20 mM H2O2 working solution is stable only for a few hours and should, therefore, be prepared fresh every time.
- The manufacturer’s protocol of the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit recommends to perform the H2O2 assay in a 100 µl total volume per reaction. We were able to successfully perform the assay with half the volume (50 µl). Therefore, the following protocol is based on a 50 µl reaction volume.
- The Amplex® Red reagent is air and light sensitive. The 1x reaction buffer is stable at room temperature. The HRP stock solution should be aliquoted and kept at -20 °C. The 20 mM H2O2 working solution is stable only for a few hours and should, therefore, be prepared fresh every time.
- Prepare an H2O2 standard curve by diluting the respective amount of 20 mM H2O2 working solution with 1x reaction buffer to obtain H2O2 standards of 0 to 10 µM, in 25 µl each. The final H2O2 concentration per reaction will be two-fold lower (e.g., 0 to 5 µM).
Notes:- The 0 µM H2O2 standard is one of the important negative controls that shows the background fluorescence coming from the mixture of Amplex® Red reagent and HRP in 1x reaction buffer in the absence of H2O2 (see negative control ‘No H2O2’ in Figure 3B).
- In our case, we used standards with the following final concentration – 0, 1, 2, 3, 4, and 5 µM H2O2 (see the loading scheme in Figure 3C).
- The 0 µM H2O2 standard is one of the important negative controls that shows the background fluorescence coming from the mixture of Amplex® Red reagent and HRP in 1x reaction buffer in the absence of H2O2 (see negative control ‘No H2O2’ in Figure 3B).
- Prepare additional negative controls. A negative control ‘Amplex® Red background’ (see control A in Figure 3B) contains only Amplex® Red reagent in 1x reaction buffer and is needed in order to monitor the background fluorescence/absorbance of the Amplex® Red reagent itself. Another negative control, termed here ‘Sample background’ (see control C in Figure 3B) is prepared for each H2O2 extract and contains the extract in 1x reaction buffer together with the HRP enzyme but without Amplex® Red reagent. This control allows to monitor the individual sample background that could come from the presence of plant compounds that could potentially serve as substrates for HRP, mimicking resorufin production.
- Pipet 25 µl of the H2O2 standards, the controls, and the samples (from step A5) into a microtiter plate.
Note: In our case, we did not have to dilute the samples with 1x reaction buffer prior to the assay. However, it may be necessary to prepare serial dilutions of the sample in order to determine the optimal sample amount per assay. As stated in the manual of the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit, high H2O2 levels may lead to lower fluorescence being detected due to the ability of the excess H2O2 to oxidize resorufin (the fluorescent reaction product) to the nonfluorescent resazurin. - Prepare a working solution of 100 µM Amplex® Red reagent and 0.2 U/ml horseradish peroxidase using the solutions from step B1 (see Table 1), in a light-protected tube (e.g., wrapped in aluminum foil).
Table 1. Working solution composition. Exemplary calculation for 2.5 ml solution, sufficient for approx. 100 assays.
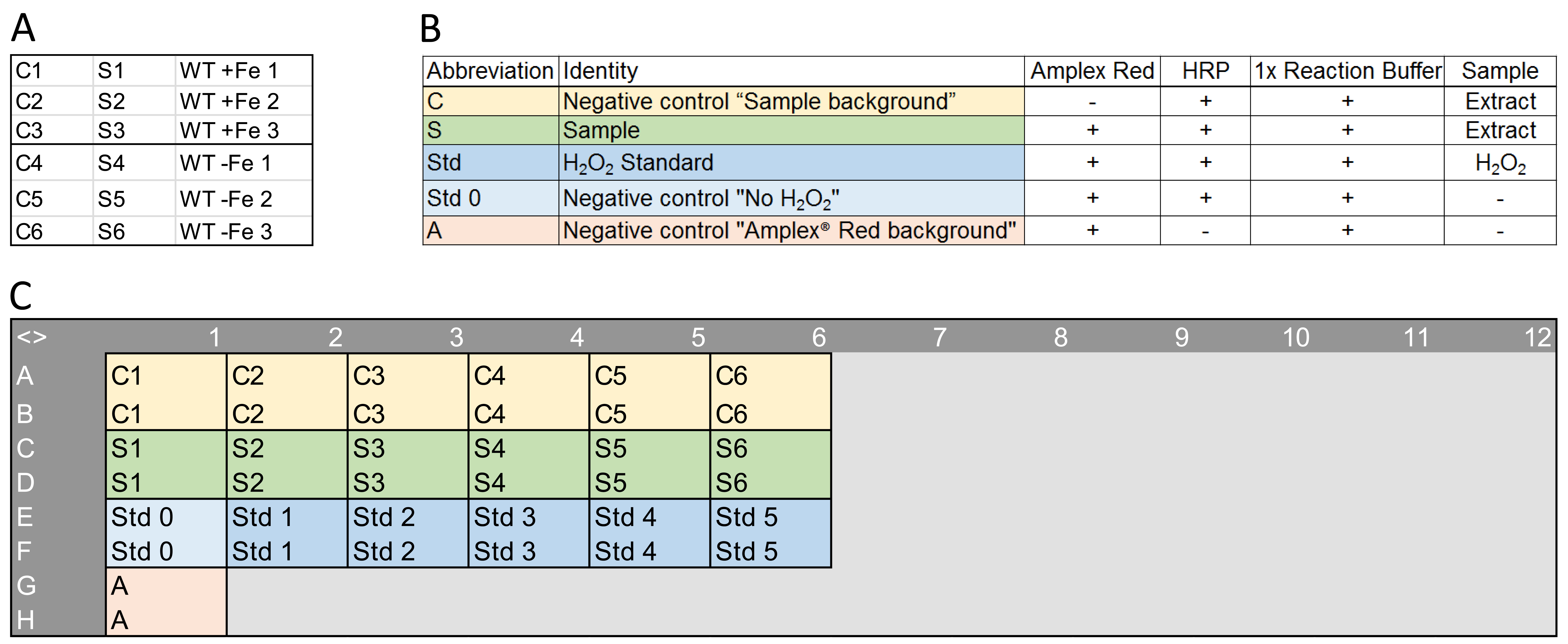
Figure 3. An example of samples, necessary controls and standards, and a plate setup. A. An example of sample identity. In this case, wild-type Arabidopsis plants (WT) were grown for 10 days on sufficient (+Fe) or deficient (-Fe) supply in three biological replicates (1 to 3). The H2O2-containing root extracts from these plants were used for the negative controls ‘Sample background’ (C1 to C6) and as samples (S1 to S6). B. A composition summary of the necessary negative controls, samples and standards. C. An example of a plate setup in a 96-well microtiter plate. Each control, sample or standard was pipetted in two technical replicates.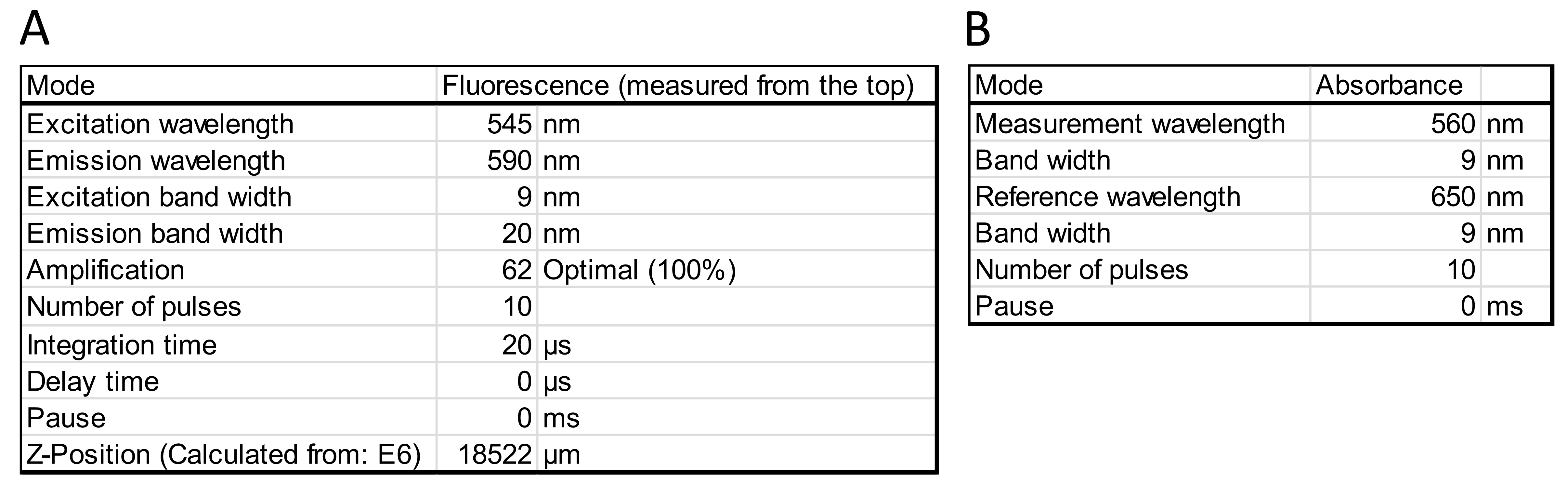
Figure 4. Example settings for the plate reader Infinite® M200 Pro for measuring resorufin fluorescence (A) and absorbance (B). Note that the Z-Position (optimal measuring height in a well) is calculated based on a well that is expected to contain a high amount of H2O2 and serves as a positive control. In this case, this is well E6 containing one of the two technical replicates of the 5 µM H2O2 standard as shown in the plate setup on Figure 3C. - Begin the reaction by adding 25 µl of working solution to each microtiter-plate well containing the H2O2 standards, the controls, and the samples (pipetted in advance in step B4). See also Note 4.
- Incubate the reactions at room temperature for 30 min in the dark.
Note: Since the assay is continuous, the reaction kinetics can be followed by performing measurements at several time points. - Detect resorufin production using a plate reader by measuring either the fluorescence (excitation at 530-560 nm, emission at 590 nm) or the absorbance at 560 nm (see Figure 4 for example settings). See Note 5.
- Prepare 10 mM Amplex® Red reagent stock solution, 1x reaction buffer (0.05 M sodium phosphate buffer, pH 7.4), 10 U/ml horseradish peroxidase (HRP) stock solution, and 20 mM H2O2 working solution according to the instructions provided with the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit.
Data analysis
We suggest to perform the H2O2 measurement on samples from three biological repetitions with two technical replicates each. Here, a technical replicate means two independent enzymatic reactions using the same control, standard or root extract.
An example of raw data, H2O2 standard curve and data analysis representation for a fluorescence measurement on root H2O2 extracts from 10 day-old seedlings is shown in Figure 5.
- Calculations based on fluorescence measurements
- Calculate the mean of the two technical replicates for the negative control ‘Amplex® Red background’, i.e., Mean (control A) (see also Figures 3B and 3C) and the mean of the two technical replicates for the H2O2 standards, i.e., Mean (Std).
- Subtract the Mean (control A) value from Mean (Std) value, i.e., Mean (Std-A).
- Correct for background fluorescence coming from the working solution components by subtracting the negative control ‘No H2O2’ (abbreviated as Std 0 in Figures 3B and 3C) from the Mean (Std-A) values for each H2O2 standard, including the Std 0 itself, so that the value for Std 0 becomes 0.
- Plot the values obtained in step A3 against the final H2O2 concentration of each standard. Obtain the equation of the trend line (see Figure 5B for an example).
- Calculate the mean fluorescence of the two technical replicates for each sample, termed Mean (Sample), and each negative control ‘Sample background’ (abbreviated as C in Figures 3B and C), termed Mean (control C).
- Correct for background fluorescence coming from the plant extract itself by subtracting the Mean (control C) values from the Mean (Sample) values, obtaining Mean (Sample-C).
- Calculate the H2O2 concentration (in µM) for each sample by using the equation obtained from the H2O2 standard curve in step A4.
- As described in the Procedure section, step A4, 200 µl phosphate buffer are added to 30 mg of tissue powder, and this ratio between tissue and buffer volume is kept throughout all samples. Therefore, 7.67 µl contain 1 mg of tissue [(200+30)/30]. Thus, 25 µl of root extract are equivalent to 3.26 mg of root material.
- The values obtained in step A7 represent the H2O2 concentration in µM (i.e., pmol/µl). Since the measurement is performed in 50 µl, multiply the number of H2O2 pmol per µl by 50 and divide this by 3.26 mg (see step A8) in order to normalize the measurement to the amount of root material per reaction. The obtained values represent the H2O2 content, e.g., in pmol H2O2/mg FW (FW-fresh weight).
- Calculate the mean H2O2 content of the three biological replicates of each line and growth condition, using the values obtained in step A9. Calculate standard deviation.
- Use the mean H2O2 content and standard deviation from step A10 for the final presentation of the data (see Figure 5C for an example).
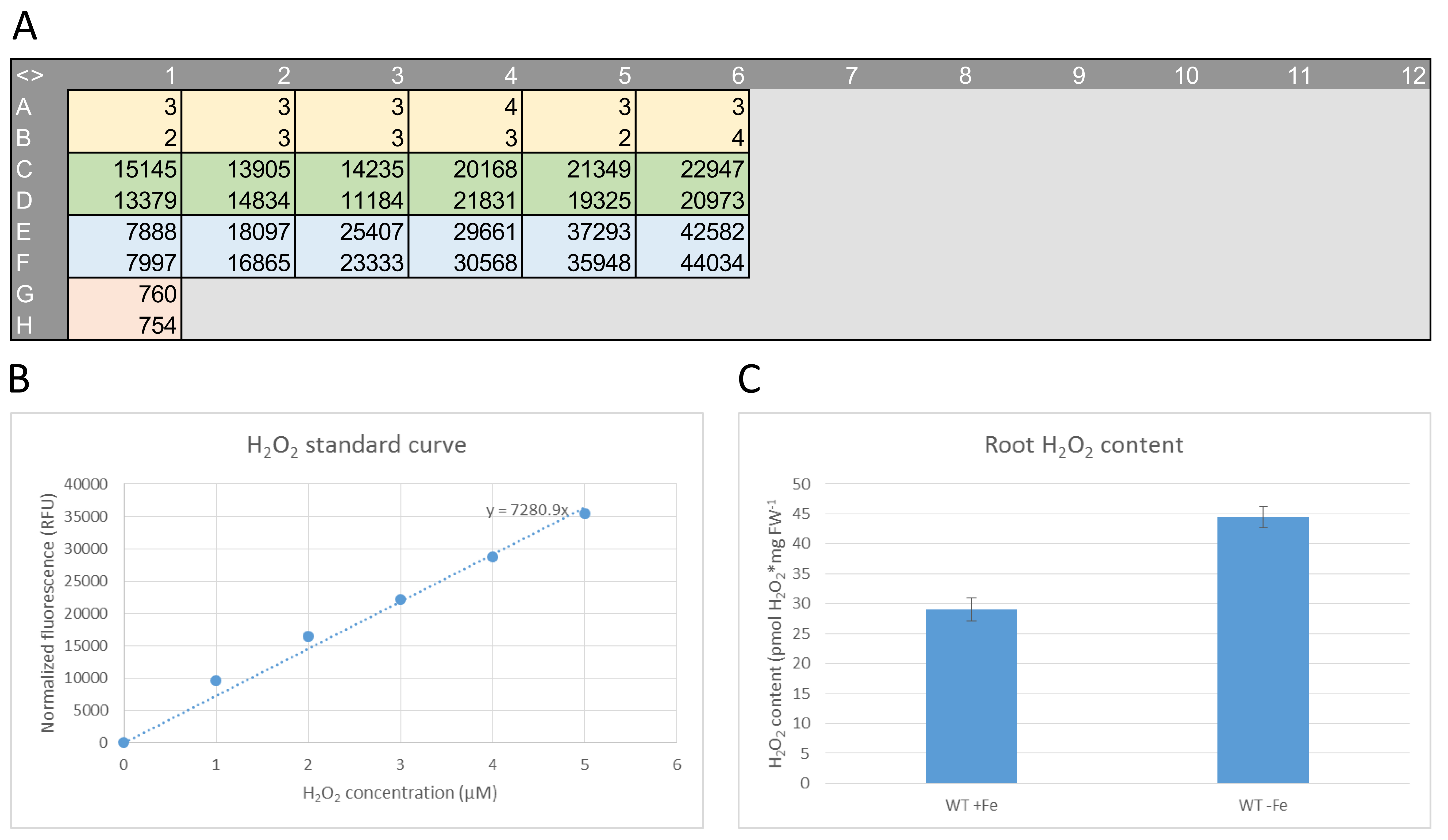
Figure 5. An example of raw data, H2O2 standard curve, and data analysis representation from a fluorescence measurement on root H2O2 extracts of 10 day-old seedlings. A. An example of raw data. The corresponding plate setup is shown in Figure 3C. B. H2O2 standard curve generated from the raw data in (A). C. An example of data analysis representation.
- Calculate the mean of the two technical replicates for the negative control ‘Amplex® Red background’, i.e., Mean (control A) (see also Figures 3B and 3C) and the mean of the two technical replicates for the H2O2 standards, i.e., Mean (Std).
- Calculations based on absorbance (Abs) measurements
- The calculation of H2O2 content based on absorbance is performed in the same way as for the fluorescence measurement. The only difference is that here two wavelengths are used – 560 nm for the measurement and 650 nm as a reference wavelength. Therefore, for the calculations, the difference between the Abs(560) and the Abs(650) values has to be used.
Notes
- Based on our experience, it is very important that the components of the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit are as fresh as possible and that they are protected from light, air and repeated freeze-thaw cycles. Suboptimal component quality results in bigger variations between the technical and biological replicates.
- Another source of variability comes from the root weight measurement. Here, it is very important to collect and freeze roots that are equally dried on paper before grinding them and determining their weight (as described in ‘Procedure’ section A1b).
- For high-throughput experiments, it may be advantageous to use a tissue homogenizer (e.g., such as Precellys® 24 homogenizer). In this case, instead of steps A2 and A3 of the ‘Procedure’ section, weigh the roots in a microcentrifuge tube, record the exact weight (e.g., ~30 mg) and grind the roots with the tissue homogenizer. Then, add the corresponding amount of ice-cold phosphate buffer, resuspend and centrifuge, as described under ‘Procedure’, steps A4 and A5.
- Since the assay is continuous, care should be taken to minimize the time difference between the first and last sample when pipetting the working solution to the samples, standards and negative controls (as described in ‘Procedure’ B6).Our preferred way is to use a multi-channel pipet when adding the working solution. In this way, the pipetting and measuring times are very similar (since the microtiter plate reader that we are using is measuring well by well).
- Often using the fluorescence measurement for calculating the H2O2 content gives less variation between the replicates, resulting in lower standard deviations and clearer differences between the different genotypes and growth conditions. However, it is advisable each time to measure both fluorescence and absorbance.
Recipes
- Sterilization solution
6% NaOCl
0.1% Triton X-100 - Hoagland medium
0.75 mM MgSO4
0.5 mM KH2PO4
1.25 mM KNO3
1.5 mM Ca(NO3)2
50 µM KCl
50 µM H3BO3
10 µM MnSO4
2 µM ZnSO4
1.5 µM CuSO4
0.075 µM (NH4)6Mo7O24
1% sucrose (pH 5.8)
1.4% plant agar
The medium was supplemented with 50 µM FeNaEDTA (sufficient Fe supply) or 0 Fe, 50 µM ferrozine (deficient Fe supply) - Phosphate buffer
20 mM K2HPO4 (pH 6.5)
Acknowledgments
This protocol was adapted from our published work (Le et al., 2016) and the manual of the Amplex® Red Hydrogen Peroxide/Peroxidase Assay Kit (https://tools.thermofisher.com/content/sfs/manuals/ mp22188.pdf). This work was supported by the Heinrich-Heine University, Düsseldorf, Germany.
References
- Le, C. T., Brumbarova, T., Ivanov, R., Stoof, C., Weber, E., Mohrbacher, J., Fink-Straube, C. and Bauer, P. (2016). ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) interacts with FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) linking iron deficiency and oxidative stress responses. Plant Physiol 170(1): 540-557.
- Lingam S1, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P. (2011). Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23(5): 1815-1829
- Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci 7(9): 405-410.
- Mittler, R. and Blumwald, E. (2015). The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27(1): 64-70.
- Xia, X. J., Zhou, Y. H., Shi, K., Zhou, J., Foyer, C. H. and Yu, J. Q. (2015). Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J Exp Bot 66(10): 2839-2856.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Brumbarova, T., Le, C. T. T. and Bauer, P. (2016). Hydrogen Peroxide Measurement in Arabidopsis Root Tissue Using Amplex Red. Bio-protocol 6(21): e1999. DOI: 10.21769/BioProtoc.1999.
- Le, C. T., Brumbarova, T., Ivanov, R., Stoof, C., Weber, E., Mohrbacher, J., Fink-Straube, C. and Bauer, P. (2016). ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) interacts with FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) linking iron deficiency and oxidative stress responses. Plant Physiol 170(1): 540-557.
Category
Plant Science > Plant biochemistry > Other compound
Plant Science > Plant metabolism > Other compound
Biochemistry > Other compound > Reactive oxygen species
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link