- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Assessment of TCR-induced Sumoylation of PKC-θ
Published: Vol 6, Iss 20, Oct 20, 2016 DOI: 10.21769/BioProtoc.1979 Views: 7555
Reviewed by: Ivan ZanoniMichael EnosMareta Ruseva

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2282 Views

Isolation and Ex Vivo Testing of CD8+ T-Cell Division and Activation Using Mouse Splenocytes
Melissa Dolan [...] John M.L. Ebos
Aug 20, 2025 3878 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 496 Views
Abstract
Sumoylation controls many cellular processes. Protein kinase C-θ (PKC-θ), a member of the Ca2+-independent PKC subfamily of kinases, serves as a regulator of T cell activation by mediating the T cell antigen receptor (TCR)- and coreceptor CD28-induced activation of the transcription factors NF-κB and AP-1 and, to a lesser extent, NFAT, and, subsequently, interleukin 2 (IL-2) production and T cell proliferation. We recently proved that TCR-induced sumoylation of PKC-θ is required for its function in T cells (Wang et al., 2015). Here we describe the method to analyze TCR-induced sumoylation of overexpressed or endogenous PKC-θ, which is carried out by immunoprecipitation of PKC-θ followed by immunoblotting with anti-SUMO1 antibody.
Keywords: TCRBackground
Like ubiquitination, sumoylation is the process of covalently modifying a target protein with SUMO. To disrupt the non-covalent interactions and to detect sumoylation specifically on the protein of interest, a stringent condition for cell lysis, immunoprecipitation and washing should be used. However, lysing cells with lysis buffer containing 1% or more SDS yields highly viscous cell lysates, making it difficult to proceed to the immunoprecipitation and immunoblotting steps. Here we describe an alternative lysing-denaturing procedure. Firstly, cells were lysed in lysis buffer supplemented with the SUMO specific proteases inhibitor N-ethylmaleimide. After centrifugation, 1% SDS was added into the supernatant of the cell lysates. The lysates were diluted 10-folds with lysis buffer supplemented with N-ethylmaleimide and subjected to immunoprecipitation. This protocol could also be adopted to detect other ubiquitin-like modification such as neddylation.
Materials and Reagents
- Corning 50 ml conical tube (Corning, catalog number: 430829 )
- 1.5 ml microcentrifuge tube (Corning, Axygen®, catalog number: MCT-150-C )
- MACS columns (Miltenyi Biotec, catalog number: 130-042-401 )
- MACS separators (Miltenyi Biotec, catalog number: 130-042-501 )
- Raji B cells (ATCC, catalog number: CCL86 )
- Jurkat T cells, Clone E6.1 (ATCC, catalog number: TIB-152 )
- Hanks’ balanced salt solution (HBSS) (Thermo Fisher Scientific, GibcoTM, catalog number: 14175095 )
- Ficoll (Tbdscience, catalog number: HY2015 )
- Phosphate-buffered saline (PBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10010023 )
- Ethylenediaminetetraacetate acid disodium salt (EDTA) (Sangon Biotech, catalog number: A610185 )
- Bovine serum albumin (BSA)
- CD4 microbeads (Miltenyi Biotec, catalog number: 130-045-101 )
- Superantigen SEE (Toxin Technology, catalog number: ET404 )
- RPMI-1640 medium (GE Healthcare, HyCloneTM, catalog number: SH30027.01 )
- Heat-inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270-106 )
- Antibiotic-antimycotic solution 100x (Thermo Fisher Scientific, GibcoTM, catalog number: 15240062 )
- N-ethylmaleimide (Sigma-Aldrich, catalog number: E1271-5G )
- Sodium dodecyl sulfate (SDS) (Sangon Biotech, catalog number: A600485 )
- Antibodies
Anti-CD3 (affymetrix, eBioscience, catalog number: 16-0037-85 )
Anti-CD28 (affymetrix, eBioscience, catalog number: 16-0289-85 )
Goat anti-mouse IgG (Jackson ImmunoResearch, catalog number: 115-001-003 )
Anti-Flag (Sigma-Aldrich, catalog number: F3165 )
Anti-PKC-θ (Santa Cruz Biotechnology, catalog number: sc-1875 ; BD, Transduction LaboratoriesTM, catalog number: 610089 )
Anti-SUMO1 (Santa Cruz Biotechnology, catalog number: sc-9060 ) - Tris (Sangon Biotech, catalog number: A600194 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S3014 )
- Nonidet P40 (Sangon Biotech, catalog number: A600385 )
- Aprotinin (EMD Millipore, catalog number: 616370 )
- Leupeptin (Calbiochem, catalog number: 108976 )
- Phenylmethanesulfonyl fluoride (PMSF) (Sigma-Aldrich, catalog number: P7626 )
- Sodium pyrophosphate tetrabasic decahydrate (NaPPi) (Sigma-Aldrich, catalog number: S6422 )
- Sodium orthovanadate (Na3VO4) (Sigma-Aldrich, catalog number: 567540 )
- Protein G sepharose (GE Healthcare, catalog number: 17-0618-01 )
- Lysis buffer (see Recipes)
Equipment
- Haemocytometer (Beckman Coulter, model: Z1 COULTER COUNTER )
- Humidified CO2 incubator (Thermo Fisher Scientific, Thermo ScientificTM, model: FormaTM 3111 )
- Laminar air flow bio-safety cabinet (ESCO Micro, model: Airstream class II )
- Centrifuge (Eppendorf, model: 5417R )
Software
- ImageJ software
Procedure
- Cell preparation and cell culture
- Dilute 500 ml blood with 500 ml HBSS, carefully layer 35 ml of diluted blood over 15 ml of Ficoll in a 50 ml conical tube.
- Centrifuge at 350 x g for 40 min at 25 °C in a swinging-bucket rotor without brake.
- Aspirate the upper layer leaving the mononuclear cell layer undisturbed.
- Carefully transfer the mononuclear cell layer to a new 50 ml conical tube. Fill the tube with HBSS, mix, and centrifuge at 300 x g for 10 min at 25 °C.
- Carefully remove supernatant. Resuspend the cell pellet in 50 ml HBSS and centrifuge at 200 x g for 15 min at 25 °C.
- Carefully remove supernatant. Resuspend cell pellet in 10 ml PBS containing 2 mM EDTA and 0.5% BSA. Determine cell number.
- Centrifuge cell suspension at 300 x g for 10 min at 25 °C. Aspirate supernatant completely.
- Resuspend cell pellet in 80 µl of PBS containing 2 mM EDTA and 0.5% BSA per 107 cells, add 20 µl of CD4 microbeads per 107 cells. Mix and incubate for 15 min in the refrigerator at 4 °C.
- Wash cells by adding 2 ml of PBS containing 2 mM EDTA and 0.5% BSA per 107 cells and centrifuge at 300 x g for 10 min at 25 °C.
- Remove the supernatant and resuspend up to 108 cells in 500 µl of PBS containing 2 mM EDTA and 0.5% BSA. Then do the magnetic separation according to the CD4 microbeads data sheet to enrich the CD4+ T cells, and save the flow through CD4- fraction of the mononuclear cells as primary antigen-presenting cells (APCs), which is contaminated with very low percentage of CD8+ T cells but not affecting the following stimulation.
- Culture the CD4+ T cells or Jurkat T cells, primary APCs or Raji B cells, in RPMI-1640 medium supplemented with 10% heat-inactivated FBS and 1% antibiotic-antimycotic solution.
- Cell stimulation
- Superantigen (SEE) stimulation
- Incubate primary APCs (1 x 107) or Raji B cells (1 x 107) with or without 500 ng/ml (primary APCs) or 100 ng/ml (Raji B) SEE in 200 μl final volume of RPMI-1640 medium (10% FBS) in a 1.5 ml microcentrifuge tube for 30 min at 37 °C. Then centrifuge the cells at 300 x g for 3 min at 25 °C and wash the pellet with ice cold PBS to obtain SEE-loaded APC-SEE or Raji B-SEE cells.
- Wash primary CD4+ or Jurkat T cells (1 x 107) with 1 ml ice cold PBS once and resuspend in 200 μl final volume of RPMI-1640 medium, and incubate on ice for 15 min.
- Mix APC-SEE or Raji-B-SEE with T cells at a 1 to 1 ratio (in 400 µl final volume), spin down at 4 °C, 300 x g, 1 min. Keep the cell pellet in the medium and immediately incubate the cell pellet for 1, 5, 15, 30 min at 37 °C.
- Antibody stimulation
- Resuspend Jurkat T (1 x 107) or human primary CD4+ cells (1 x 107) in 200 μl of serum free RPMI. Incubate on ice for 10 min. Then add 10 μg/ml anti-CD3 and/or 2 μg/ml anti-CD28 mAbs, place the mixture on ice for 5 min, followed by crosslinking with goat-anti-mouse IgG (10 μg/ml) and immediately incubate the cells at 37 °C for 1, 5, 15, 30 min .
- Centrifuge the cells at 350 x g for 3 min, carefully remove the supernatant and lyse stimulated cells in 120 μl lysis buffer (Xie et al., 2013) containing 20 mM N-ethylmaleimide (NEM) on ice for 10 min.
- Clear the lysate by centrifugation (8,500 x g, 10 min, 4 °C), then add 1% SDS (v/v) to the supernatants and dissociate the immunoprecipitated proteins by heating at 90 °C for 10 min.
- Dilute lysate obtained in step 4 ten-folds with lysis buffer, and immunoprecipitate Flag-tagged PKC-θ or endogenous PKC-θ with anti-Flag M2 or goat-anti-PKC-θ Abs, respectively, at 4 °C overnight with rotation. Anti-IgG immunoprecipitation and IPs from lysates of unstimulated cells are done in parallel as negative controls. Then add 30 µl protein G beads (50% [v/v] in PBS) and incubate for 2-4 h at 4 °C with rotation.
- Centrifuge at 8,000 x g for 5 min at 4 °C, remove the supernatant carefully and wash the immunoprecipitates extensively by adding 1 ml lysis buffer containing 20 mM NEM and vortexing for 3-5 min.
- Repeat the wash step for another 4 times. Then subject the samples to SDS-PAGE, and immunoblot with anti-SUMO1 (1:1,000) or mouse-anti-PKC-θ (1:1,000) Abs. A representative immunoblotting is shown in the Representative data section (Figure 5).
Data analysis
- To exclude the non-specific bands detected by the anti-SUMO1 antibody, a set of negative controls including anti-IgG immunoprecipitates (IPs) and IPs from lysates of unstimulated cells should be included in the pilot experiment. The system works if the signal of the control lanes is barely detectable and the cell stimulation is successful.
- Analyze signals with ImageJ software as followed:
Click ‘File’ and ‘Open’, then choose the image. - Click the ‘Rectangular’ tool, a box with yellow lines appears. Drag around the signals to define the area to be analyzed. Click ‘Ctrl+1’ to select the first line (Figure 1).
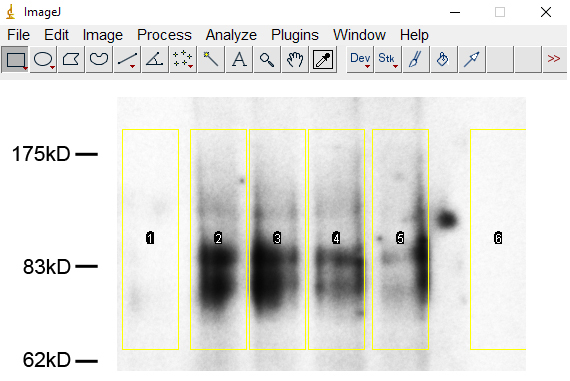
Figure 1. Selection of the sumoylated bands of PKC-θ in ImageJ - Move the box with arrow keys to next lane without changing the size of the box, click ‘Ctrl+2’ to select next lane. Repeat this step till all lanes are selected.
- Move the box with arrow keys to a blank area, click ‘Ctrl+2’ to select the background.
- Click ‘Ctrl+3’ to plot lanes. A series of plots will appear in a new window (Figure 2).
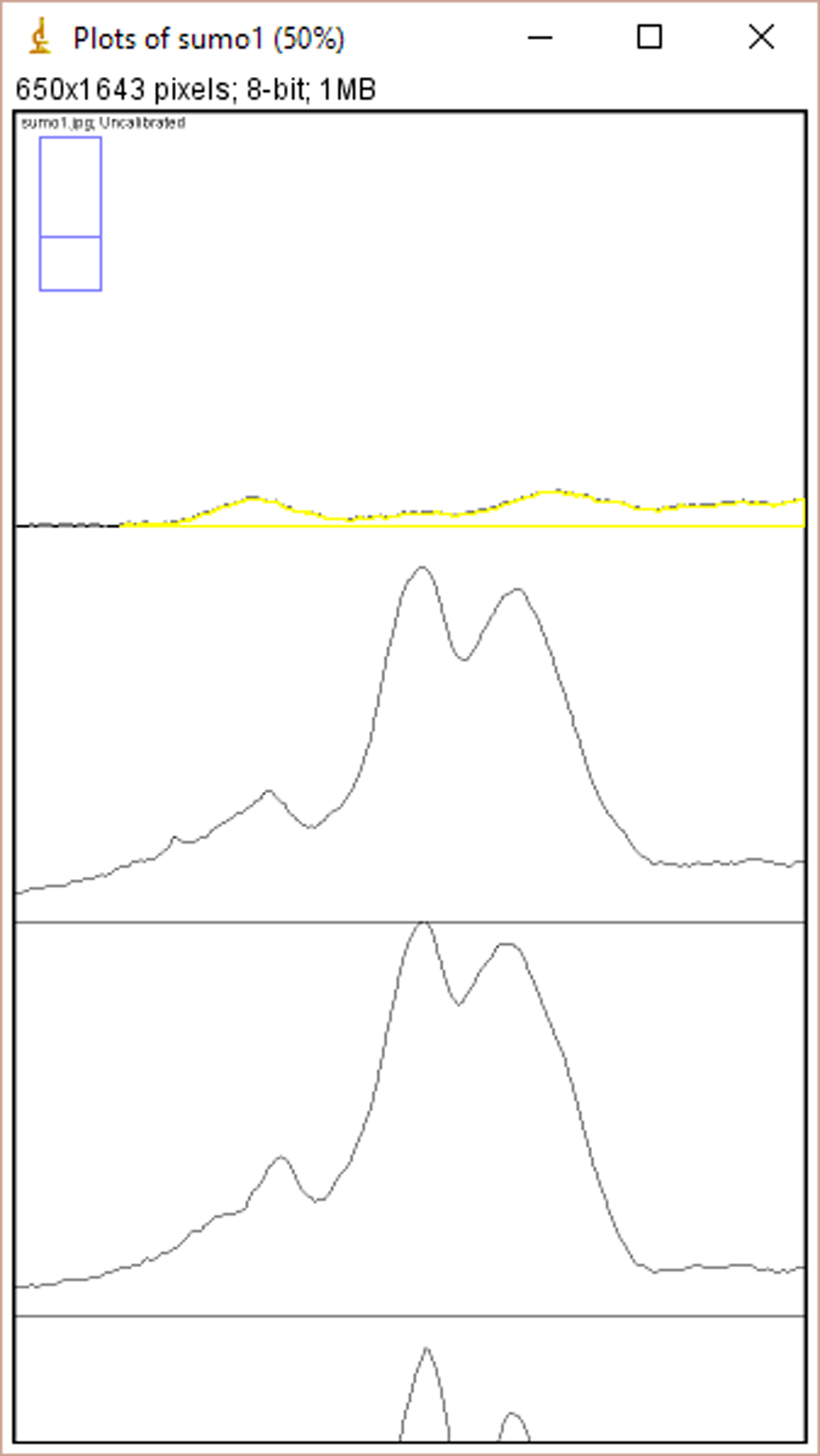
Figure 2. The window of ImageJ after Clicking ‘Ctrl+3’ - Select ‘Wand tool’ and click the area of each plot (Figure 3). A new window opens with the volume data representing the pixel intensity inside a defined boundary. Subtract background pixel intensity (using value of blank area) from all values to obtain the relative intensity values (Figure 4).
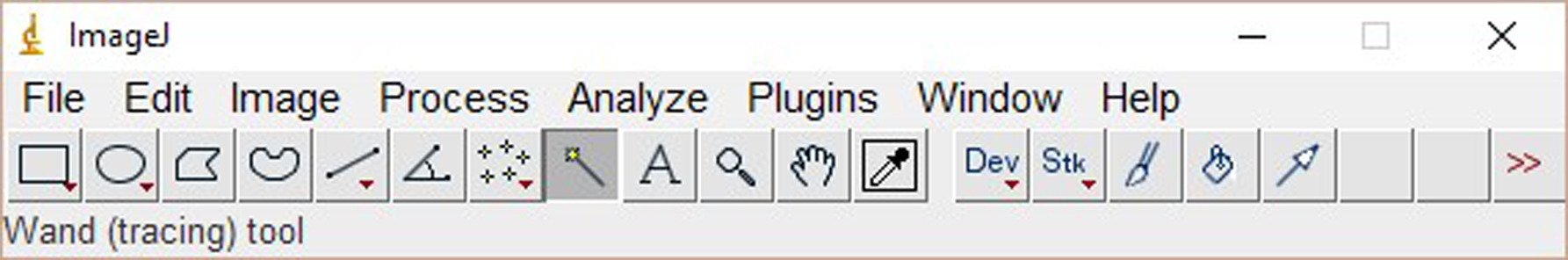
Figure 3. Selection of the ‘Wand tool’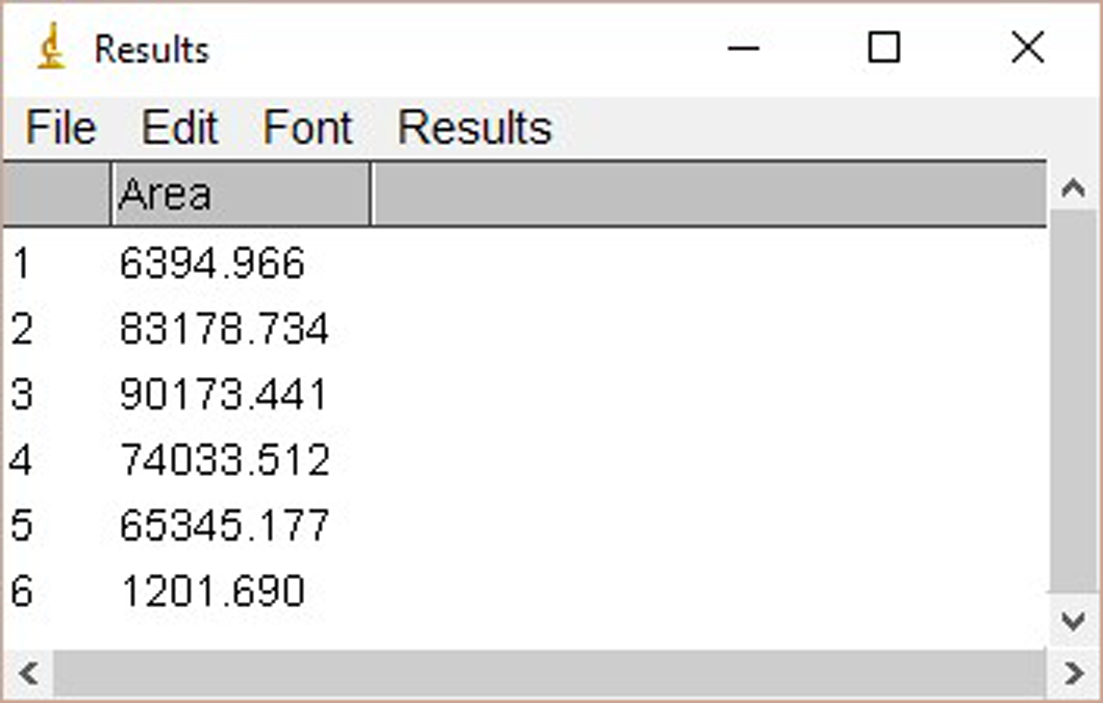
Figure 4. The interface of ImageJ after analysis - Quantify signals of PKC-θ blot in the similar way.
- Normalize the SUMO1 value of each lane with its corresponding PKC-θ value.
Representative data
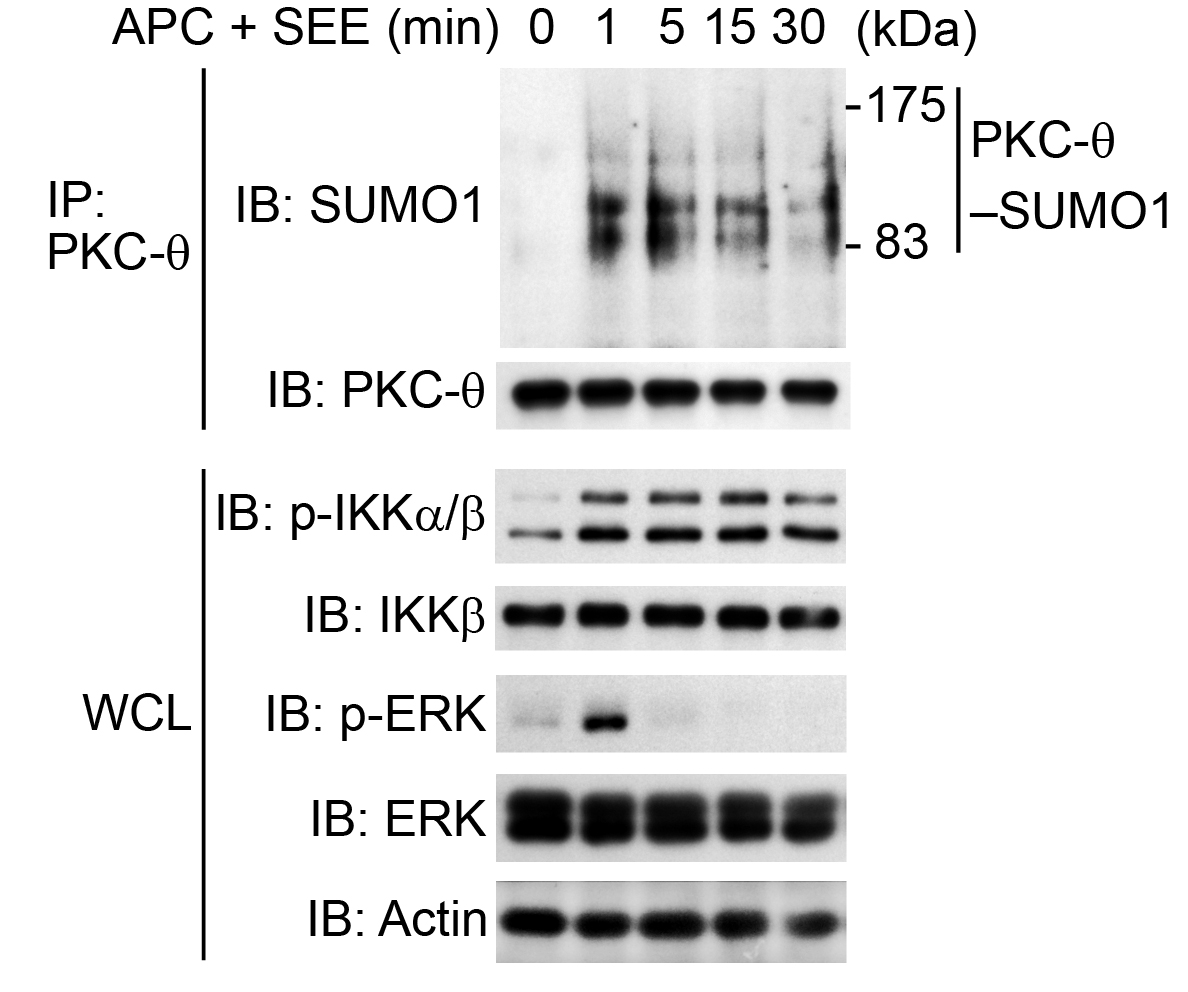
Figure 5. In vivo assay of the sumoylation of PKC-θ (PKC-θ-SUMO1) in human primary CD4+ T cells. Freshly isolated human primary CD4+ T cells were stimulated for 0-30 min (above lanes) with SEE-pulsed human APCs (APC + SEE), at a ratio of 1:1, assessed by immunoblot analysis (IB) of proteins immunoprecipitated (IP) with anti-PKC-θ (top) and of whole-cell lysates (WCL) without immunoprecipitation (bottom), probed with antibodies to various molecules (left margin); actin serves as a loading control (Wang et al., 2015).
Notes
- Larger apparent molecular size of the protein should be anticipated due to multiple SUMO attachment sites. Thus a gel with a lower concentration of acrylamide (i.e., 8%) is recommended.
- It is important that cells should be lysed as quickly as possible after stimulation, since sumoylation often occurs transiently and is easily deconjugated by the SUMO-specific proteases. Moreover, 20 mM of N-ethylmaleimide, a chemical inhibitor of these proteases is recommended to be supplemented in the lysis buffer in both the lysing and washing steps during the immunoprecipitation procedure.
- Since the signal of the sumoylated bands of endogenous PKC-θ could be very weak, overnight incubation of the membrane with anti-SUMO1 Ab at 4 °C and longer exposure time when visualizing the immunoblot signals are highly recommended.
Recipes
- Lysis buffer
20 mM Tris–HCl (pH 7.5)
150 mM NaCl
5 mM EDTA
1% Nonidet P40
10 μg/ml aprotinin
10 μg/ml leupeptin
1 mM PMSF
5 mM NaPPi
1 mM Na3VO4
Acknowledgments
This protocol is modified from the in vivo ubiquitination assay (Wertz et al., 2004). Supported by the National Natural Science Foundation of China (31170846) and the Ministry of Science and Technology of China (2013CB835300).
References
- Wang, X. D., Gong, Y., Chen, Z. L., Gong, B. N., Xie, J. J., Zhong, C. Q., Wang, Q. L., Diao, L. H., Xu, A., Han, J., Altman, A. and Li, Y. (2015). TCR-induced sumoylation of the kinase PKC-θ controls T cell synapse organization and T cell activation. Nat Immunol 16(11): 1195-1203.
- Wertz, I. E., O'Rourke, K. M., Zhou, H., Eby, M., Aravind, L., Seshagiri, S., Wu, P., Wiesmann, C., Baker, R., Boone, D. L., Ma, A., Koonin, E. V. and Dixit, V. M. (2004). De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 430(7000): 694-699.
- Xie, J. J., Liang, J. Q., Diao, L. H., Altman, A. and Li, Y. (2013). TNFR-associated factor 6 regulates TCR signaling via interaction with and modification of LAT adapter. J Immunol 190(8): 4027-4036.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Wang, X., Chen, Z., Wang, Q. and Li, Y. (2016). Assessment of TCR-induced Sumoylation of PKC-θ. Bio-protocol 6(20): e1979. DOI: 10.21769/BioProtoc.1979.
Category
Immunology > Immune cell function > Antigen-specific response
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









