- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Spectrophotometric Determination of Glutamine Synthetase Activity in Cultured Cells
Published: Vol 6, Iss 19, Oct 5, 2016 DOI: 10.21769/BioProtoc.1959 Views: 13951
Reviewed by: Masahiro MoritaRaghuveer KavarthapuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Preparation of Testicular Cells for Immunofluorescence Analysis of Manchette in Elongating Spermatids
Changmin Niu [...] Zhibing Zhang
Jun 20, 2025 2386 Views

Fluorescence Polarization-Based High-Throughput Screening Assay for Inhibitors Targeting Cathepsin L
Keyu Guo [...] Shuyi Si
Jul 20, 2025 2282 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 496 Views
Abstract
Glutamine synthetase (GS), which catalyzes the conversion of glutamate and ammonia to glutamine, is widely distributed in animal tissues and cell culture lines. The importance of this enzyme is suggested by the fact that glutamine, the product of GS-catalyzed de novo synthesis reaction, is the most abundant free amino acid in blood (Smith and Wilmore, 1990). Glutamine is involved in many biological processes including serving as the nitrogen donor for biosynthesis, as an exchanger for the import of essential amino acids, as a means to detoxifying intracellular ammonia and glutamate, and as a bioenergetics nutrient to fuel the tricarboxylic acid (TCA) cycle (Bott et al., 2015). The method for the assay of GS enzymatic activity relies on its γ-glutamyl transferase reaction by measuring γ-glutamylhydroxamate synthesized from glutamine and hydroxylamine, and the chromatographic separation of the reaction product from the reactants (Deuel et al., 1978). An overview of the GS glutamyl transferase reaction can be found in Figure 1. GS activity was measured by a spectrophotometric assay at a specific wavelength of 560 nm using a microplate reader. The method is simple, and has a comparable sensitivity with those methods applying radioactively labelled substrates. This modified procedure has been applied to assay/determine GS activity in cultured cell lines including the human mammary epithelial MCF10A cells and the murine pre-B FL5.12 cells, and could be used to measure GS activity in other cell lines.
Figure 1. An overview of the GS glutamyl transferase reaction
Materials and Reagents
- 0.05% trypsin-EDTA (1x) (Thermo Fisher Scientific, GibcoTM, catalog number: 25300-054 )
- PBS (1x) (Corning, catalog number: 21-031-CV )
- Imidazole (Sigma-Aldrich, catalog number: I0250 )
- L-glutamine (Sigma-Aldrich, catalog number: G3126 )
- Hydroxylamine hydrochloride (Sigma-Aldrich, catalog number: 55459 )
- Sodium arsenate dibasic heptahydrate (Sigma-Aldrich, catalog number: S9663 )
- Manganese(II) chloride tetrahydrate (MnCl2·4H2O) (Sigma-Aldrich, catalog number: 221279 )
- Adenosine 5’-diphosphate sodium salt (ADP) (Sigma-Aldrich, catalog number: A2754 )
- Iron(III) chloride hexahydrate (FeCl3) (Sigma-Aldrich, catalog number: 31232 )
- Trichloroacetic acid (Sigma-Aldrich, catalog number: 522082 )
Note: This product has been discontinued. - Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 258148 )
- L-glutamic acid γ-monohydroxamate (γ-glutamylhydroxamate) (Sigma-Aldrich, catalog number: G2253 )
- Lysis buffer (see Recipes)
- 1x assay buffer (see Recipes)
- 1x stop buffer (see Recipes)
- γ-glutamylhydroxamate standard stock (see Recipes)
Equipment
- AccumetTM AB15 Basic and BioBasicTM pH/mV/°C Meters (Thermo Fisher Scientific, Fisher Scientific, catalog number: 13-636-AB15B )
- Sonicator (model: CL-18/120 ) (Thermo Fisher Scientific, Fisher ScientificTM, catalog number: FB120110 )
- -80 °C freezer
- 37 °C incubator
- Microcentrifuge (Eppendorf, model: 5418 )
- Microplate (96-well plate clear bottom) reader (Molecular Devices, model: SpectraMax M5 )
Software
- SoftMaxPro software
Procedure
- Collect ~8 x 106 cells in a 10 cm dish cells by using 1x trypsin.
- Rinse the cells with cold 1x PBS and then remove the PBS twice.
- Add certain amount (i.e., 300-500 μl) of lysis buffer (50 mM imidazole-HCl, pH 6.8) and lyse the cells with sonicator at level 4 output control, 50% duty cycle for 30 sec and pause for 30 sec for 5 cycles.
- Incubate the cells at -80 °C for at least 4 h for the freeze/thaw cell lysis procedure.
- Thaw the cells and centrifuge the cells at 16,000 x g at 4 °C for 15 min.
- Collect the supernatant and measure the protein concentration by BCA protein assay (He, 2011).
- Add 20-40 μg of protein, then add lysis buffer to raise volume to 50 μl.
- Add equal volume (50 μl) of 1x assay buffer and then incubate at 37 °C for 2~6 h (Please see the notes).
- Add equal volume (100 μl) of 1x stop buffer to observe brown color products and centrifuge at 16,000 x g at 4 °C for 10 min.
- The absorbance of γ-glutamylhydroxamate in the supernatant of samples was measured at 560 nm. The blank was 50 μl lysis buffer + 50 μl 1x assay buffer + 100 μl 1x stop buffer.
- A serial dilution of 25, 12.5, 6.25, 3.125, 1.5623, 0.781, and 0.391 mM γ-glutamylhydroxamate (100 μl) were added with 100 μl of 1x stop buffer and centrifuge at 16,000 x g at 4 °C for 10 min. The absorbance of γ-glutamylhydroxamate in the supernatant was measured at 560 nm at room temperature as a standard curve (Figure 2).
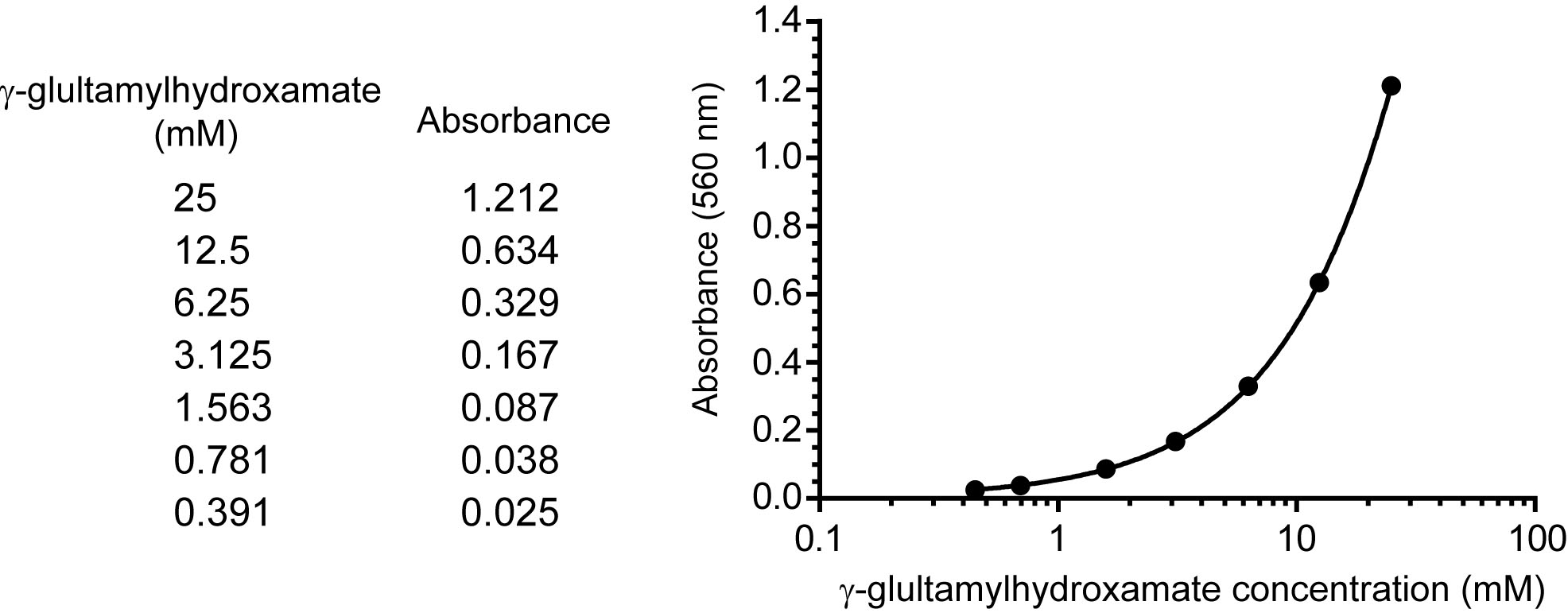
Figure 2. A representative standard curve. The standard curve is generated by performing a serial dilution of γ-glutamylhydroxamate. The upper boundary of the standard starts at a known concentration of 25 mM, and this is diluted by half to reach the lower boundary at 0.39 mM. A blank with no standard is included. These concentrations are then input into SoftMaxPro software and the absorbance is read at 560 nm. The readings from known concentrations are determined and a 4 parameter fit is applied. In this way the standard curve is generated within a large working range, unknown samples can be tested, and the absorbance of unknowns can be plotted to determine concentration.
Representative data
The GS activity was reported as nmol min-1 g protein-1. The GS activity was expressed by the formula: γ-glutamylhydroxamate concentration (mM) x 100 μl/incubation time (min)/protein amount (g). For example, the concentration of γ-glutamylhydroxamate calculated from MCF10A vector cells was 2.378 mM as compared with standard curve. The added protein amount was 10.36 μg, and the incubation time at 37 °C was 120 min. The GS activity would be 2.378 x 100/120/10.36 x10-6 = 1.91 x 105 nmol min-1 g protein-1 as indicated (Figure 3).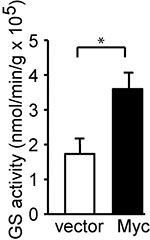
Figure 3. Myc induces GS activity. c-Myc was stably expressed in MCF10A cells. GS activity was determined and is shown as the mean plus SEM of at least five independent experiments. *P < 0.05. Student’s t-test was used to determine the P value (Bott et al., 2015).
Notes
The various incubation time (2-6 h) at 37 °C was determined by the amount of active GS expressed in different cell lines. Higher active GS expressed leads to shorter incubation time to reach the optimal amount of γ-glutamylhydroxamate. The optimal incubation time used in the assay may be adjusted for different cell lines.
Recipes
- Lysis buffer (50 mM imidazole-HCl, pH 6.8)
Add 170.2 mg imidazole in 50 ml ddH2O and adjust pH to 6.8 with HCl. - 1x assay buffer
- 5x stock 250 mM imidazole-HCl, pH 6.8
Add 851 mg imidazole in 50 ml ddH2O and adjust pH to 6.8 with HCl. - 5x stock 250 mM L-glutamine, 250 mM imidazole-HCl
Add 1.8275 g L-glutamine in 50 ml 5x stock 250 mM imidazole-HCl. - 5x stock 125 mM hydroxylamine
Add 434.3 mg hydroxylamine in 50 ml ddH2O. - 5x stock 125 mM sodium arsenate
Add 1.95 g sodium arsenate in 50 ml ddH2O. - 5x stock 10 mM MnCl2
Add 99.2 mg MnCl2 in 50 ml ddH2O. - 5x stock 0.8 mM ADP
Add 17.09 mg ADP in 50 ml ddH2O.
- 5x stock 250 mM imidazole-HCl, pH 6.8
- 1x stop buffer
3x stop buffer (270 mM FeCl3, 5.4 N HCl, 4.35% trichloroacetic acid)
Use 7.3 g FeCl3, 45 ml 12 N HCl as well as 4.35 ml 100% trichloroacetic acid and add ddH2O to 100 ml to get 3x stop buffer.
Dilute 3x stop buffer with ddH2O to get 1x stop buffer (final concentration: 90 mM FeCl3, 1.8 N HCl and 1.45% trichloroacetic acid) when doing the assay. - 100 mM γ-glutamylhydroxamate standard stock
Add 16.214 mg γ-glutamylhydroxamate in 1 ml ddH2O.
Acknowledgments
This protocol was adapted and modified from a previous study (Deuel et al., 1978). It was described in (Bott et al., 2015). This work was supported by grants from NIH (R01CA129536 and R01GM97355 to WXZ).
References
- Bott, A. J., Peng, I. C., Fan, Y., Faubert, B., Zhao, L., Li, J., Neidler, S., Sun, Y., Jaber, N., Krokowski, D., Lu, W., Pan, J. A., Powers, S., Rabinowitz, J., Hatzoglou, M., Murphy, D. J., Jones, R., Wu, S., Girnun, G. and Zong, W. X. (2015). Oncogenic Myc induces expression of glutamine synthetase through promoter demethylation. Cell Metab 22(6): 1068-1077.
- Deuel, T. F., Louie, M. and Lerner, A. (1978). Glutamine synthetase from rat liver. Purification, properties, and preparation of specific antisera. J Biol Chem 253(17): 6111-6118.
- He, F. (2011). BCA (bicinchoninic acid) protein assay. Bio-protocol 101: e44.
- Smith, R. J. and Wilmore, D. W. (1990). Glutamine nutrition and requirements. JPEN J Parenter Enteral Nutr 14(4 Suppl): 94S-99S.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Peng, I., Bott, A. J. and Zong, W. (2016). Spectrophotometric Determination of Glutamine Synthetase Activity in Cultured Cells. Bio-protocol 6(19): e1959. DOI: 10.21769/BioProtoc.1959.
Category
Biochemistry > Protein > Activity
Cancer Biology > Cellular energetics > Biochemical assays
Molecular Biology > Protein > Detection
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link