- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro Assay for Dendritic Spine Retraction of Hippocampal Neurons with Sparse Labeling
Published: Vol 6, Iss 18, Sep 20, 2016 DOI: 10.21769/BioProtoc.1937 Views: 10143
Reviewed by: Soyun KimShai BerlinEmmanuelle Berret
Abstract
Dendritic spines are the post-synaptic structures that play a central role in excitatory synaptic transmission. Developmental spinogenesis relies on a variety of stimuli such as those derived from cell-cell communication and their downstream signaling. Here, we describe an in vitro assay of dendritic spine retraction using hippocampal slice culture, in which individual neurons are sparsely and brightly labeled by the Supernova method, for the study of molecular mechanisms of spine development.
Keywords: SpineMaterials and Reagents
- Materials
- Electrode (5 mm Φ platinum disk) (Nepa Gene, model: CUY650P5 )
- Surgical needle (ELP, model: CR13-50 )
- Glass capillary (Warner Instruments, model: GC150TF-10 )
- NuncTM culture plate (6-well) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 140675 )
- Millicell culture plate inserts (EDM Millipore, catalog number: PICM03050 )
- Confetti (LCR membrane filter) (EDM Millipore, catalog number: FHLC01300 )
- Petri dish (60 mm) (Sigma-Aldrich, catalog number: Z721034 )
Note: This product has been discontinued. - Microcentrifuge tube (Sigma-Aldrich, catalog number: Z666505 )
- Cover glass (MATSUNAMI GLASS)
- Microscope glass slide (MATSUNAMI GLASS)
- Aspirator tube assembly (Drummond Scientific, catalog number: 2-000-000 )
- Electrode (5 mm Φ platinum disk) (Nepa Gene, model: CUY650P5 )
- Sparse and bright labeling of hippocampal neurons using the in utero electroporation-based Supernova system (Mizuno et al., 2014)
- Pregnant mouse (E13.5-E15.5)
- Maxi-prep kit
- Trypan blue solution (0.4%) (Sigma-Aldrich, catalog number: T8154 )
- Supernova vector DNA solution (pK036.TRE-Flpe-WPRE-pA and pK037.CAG-FRT-STOP-FRT-TurboRFP-ires-tTA-WPRE-pA)
- Somnopentyl (Pentobarbital sodium)
- 70% ethanol
- Saline
- Pregnant mouse (E13.5-E15.5)
- Organotypic hippocampal slice culture
- Mouse pup (P4-P5)
- MEM with gluramax-1 (Thermo Fisher Scientific, catalog number: 41090-028 )
- EBSS (Thermo Fisher Scientific, catalog number: 14155-048 )
- D-glucose (NACALAI TESQUE, catalog number: 16805-35 )
- Penicillin-streptomycin (Thermo Fisher Scientific, GibcoTM, catalog number: 15140-122 )
- Nystatin (Thermo Fisher Scientific, GibcoTM, catalog number: 15340029 )
- HEPES (1 M) (Thermo Fisher Scientific, GibcoTM, catalog number: 15630-080 )
- Horse serum (Sigma-Aldrich, catalog number: H1270-500 ml )
- Culture medium (See Recipes)
- Slicing buffer(See Recipes)
- Mouse pup (P4-P5)
- In vitro retraction assay
- EphrinA3-Fc (R&D Systems, catalog number: BT359 )
- Human Fc fragment (Jackson ImmunoResearch, catalog number: 009-000-008 )
- Goat anti-human IgG, Fc fragment specific (Jackson ImmunoReserach, catalog number: 109-001-008 )
- Paraformaldehyde (NACALAI TESQUE, catalog number: 02890-45 )
- PBS (-)
- Vectashield mounting media with DAPI (Vector laboratories, catalog number: H-1200 )
- EphrinA3-Fc (R&D Systems, catalog number: BT359 )
Equipment
- Sparse and bright labeling of hippocampal neurons using the in utero electroporation-based Supernova system.
- Electroporator (Nepa Gene, model: Nepa21 )
- Water bath
- Puller (NARISHIGE Group, model: PC-10 )
- Heating table (Leica Biosystems, model: HI1220 )
- Surgical tools (scissor, 12 cm) (Fine Science Tools, catalog number: 14001-12 )
- Surgical tools (scissor, 10.5 cm) (Fine Science Tools, catalog number: 14088-10 )
- Surgical tools (forceps, 12 cm) (Fine Science Tools, catalog number: 11000-12 )
- Surgical tools (forceps, 10 cm) (Fine Science Tools, catalog number: 11050-10 )
- Surgical tools (ring forceps, 8.5 cm) (Fine Science Tools, catalog number: 11101-09 )
- Electroporator (Nepa Gene, model: Nepa21 )
- Organotypic hippocampal slice culture
- LinearSlicer (Dosaka, model: PRO7 )
- 37 °C, 5% CO2 incubator
- Pipette
- LinearSlicer (Dosaka, model: PRO7 )
- In vitro retraction assay
- 25 °C incubator
- 25 °C incubator
- Imaging and analysis
- Confocal microscope (Leica Biosystems, model: TSC-SP5 )
- Imaris filament tracer (Bitplane)
- Confocal microscope (Leica Biosystems, model: TSC-SP5 )
Software
- Imaris
Procedure
- Sparse and bright labeling of hippocampal neurons using the in utero electroporation-based Supernova system
Note: Using the Supernova system that enables sparse and bright cell labeling with little background is highly recommended for analyses of fine structures such as spines of individual neurons. The Supernova system is described elsewhere (Mizuno et al., 2014). Briefly, this system uses two vectors, TRE-Flpe-WPRE-pA (pK036) and CAG-FRT-STOP-FRT-TurboRFP-ires-tTA-WPRE-pA (pK037). In a small population of neurons that carry both vectors, TRE leakage drives weak but over-threshold Flpe expression from the fist vector, which is followed by removal of the FRT-STOP-FRT cassette in a few copies of the second vector and weak expression of tTA from these copies. Then, only in these sparse neurons, tTA binds with TRE and induces strong Flpe expression, which results in removal of the FRT-STOP-FRT cassette from many copies of the second vector, and finally results in extremely strong RFP expression via the positive feedback. Flpe-based Supernova vectors [TRE-Flpe-WPRE-pA (pK036) and CAG-FRT-STOP-FRT-TurboRFP-ires-tTA-WPRE-pA (pK037)] are available from T.I. (tiwasato@nig.ac.jp).- To prepare micropipettes, pull glass capillary using a puller under the following conditions: two-step; heater, 65-75; weight, 1-3. Cut off the tip with forceps for appropriate diameter (about 20 μm).
- To prepare DNA solution for the Supernova labeling, purify the plasmid set, TRE-Flpe-WPRE-pA (pK036) and CAG-FRT-STOP-FRT-TurboRFP-ires-tTA-WPRE-pA (pK037), using endotoxin free Maxi-prep kit, and mix them to a final concentration of 1 μg/μl of pK036 and 50 ng/μl of pK037.
- Aliquot 40 μl of the DNA solution, and add 4 μl of Trypan blue solution (0.4%).
- Anesthetize a pregnant mouse (E13.5-E15.5) with pentobarbital sodium through intraperitoneal injection (50 mg/kg in saline).
- Place the anesthetized mouse on a working plate, and wash the abdomen with 70% ethanol.
- Make an incision at the abdominal midline using scissors, and take out the uterus gently onto the saline-moistened gauze. During the surgery, keep the uterus moist with pre-warmed saline (Figure 1).

Figure 1. An anesthetized pregnant mouse with embryos (E14.5) - Inject 1-2 μl of the DNA solution into the target ventricle of the embryos using the micropipette with an aspirator tube.
- Hold the embryos with electrodes (Figure 2: negative electrode on the injected side of the embryo head), and apply electric pulses [40-50 V, P on 50 msec, P off 950 msec (1 Hz), 5 times].
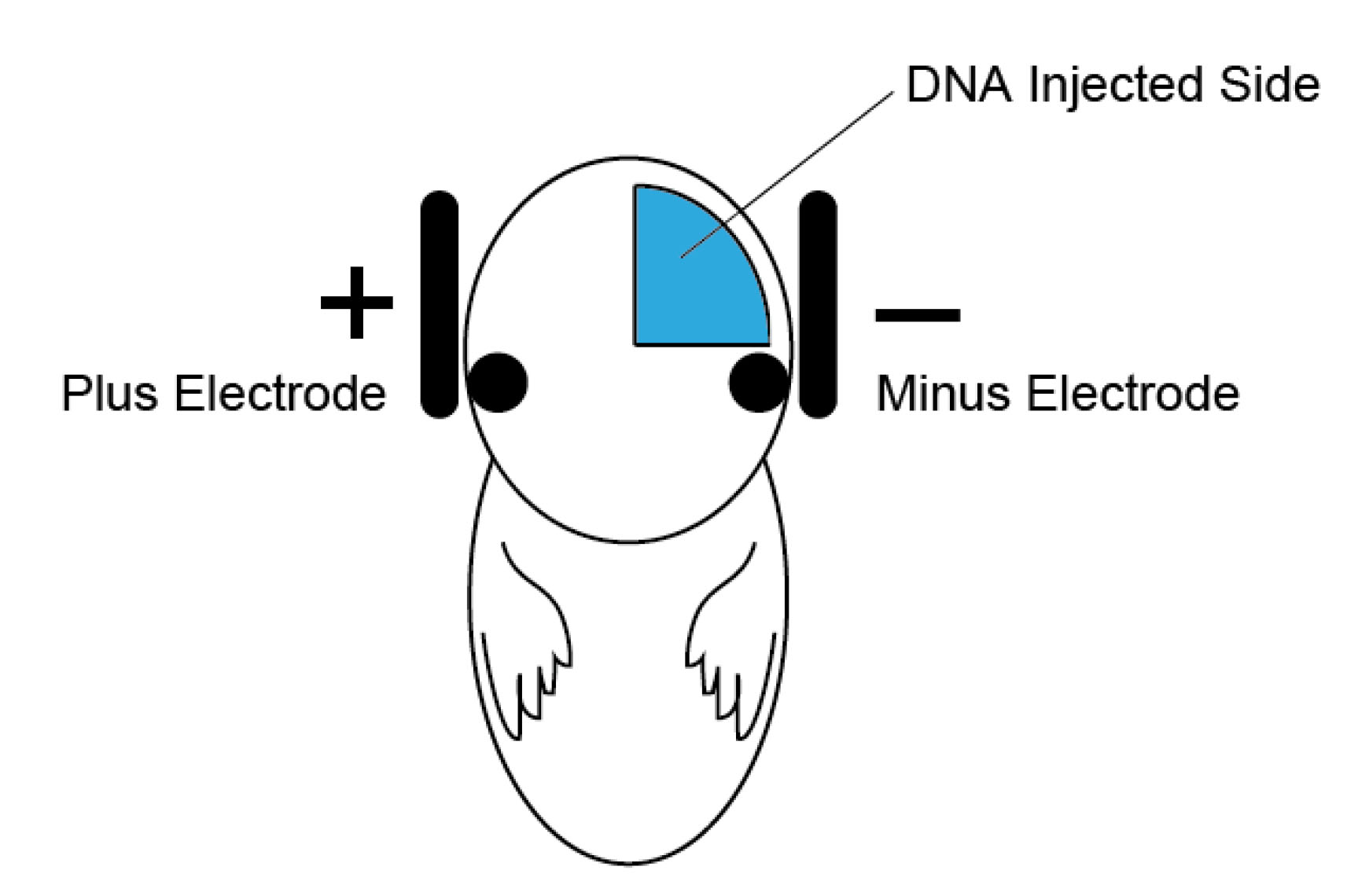
Figure 2. Schematic illustration of in utero electroporation. DNA injection side (blue) and the relative position of the electrodes for the hippocampal electroporation. - Place the uterus gently into the abdominal cavity.
- Fill the cavity with pre-warmed saline.
- Suture the surgical incision using surgical needle and thread.
- Place the mouse on a heating table at 37 °C until recovery from anesthesia.
- To prepare micropipettes, pull glass capillary using a puller under the following conditions: two-step; heater, 65-75; weight, 1-3. Cut off the tip with forceps for appropriate diameter (about 20 μm).
- Organotypic hippocampal slice culture
- Pour 1 ml of culture medium into each well (6-well plate).
- Place a Millicell culture plate insert into each well.
- Cut confetti disc using scissors (quarter), and put a piece on each insert.
- Place the plate into a 5% CO2 incubator at 37 °C.
- Pour 10 ml of slicing buffer into a Falcon tube (50 ml), and keep it on ice.
- Pour 5 ml of slicing buffer into a Petri dish (60 mm) on ice.
- Euthanize pup (P4-P5) by decapitation using scissors.
- Place the brain into a Petri dish containing slicing buffer on ice.
- Cut the brain half along the midline after removing the cerebellum.
- Glue the brain tissue on the stage of LinearSlicer buffer tray.
- Fill the buffer tray with the slicing buffer (4 °C).
- Cut a 300 μm hippocampal slices (sagittal sections) using LinearSlicer. Leave the entorhinal cortex attached to the hippocampus (Figure 3).
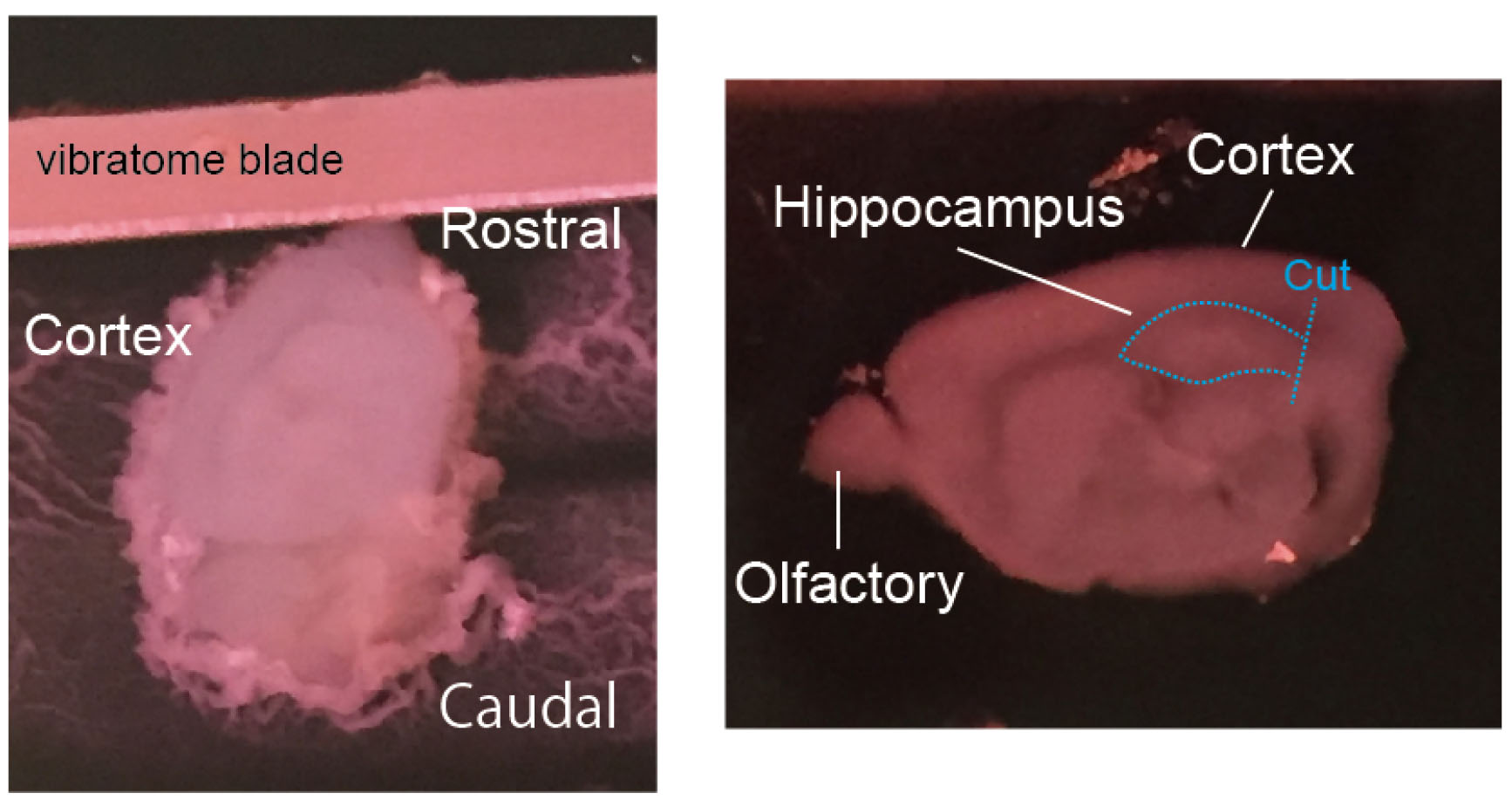
Figure 3. Dissection of the hippocampus from the brain. Left: A P5 mouse brain was sagittally sliced by a vibratome. Right: A 300 μm-thick slice and cut position were shown. - Collect the slices in a 50 ml Falcone tube containing slicing buffer (4 °C).
- Take a slice from the tube using a pipette, and put it on the center of the confetti on insert.
- Remove excess buffer from the insert, and place the plate in a 5% CO2 incubator (37 °C).
- Change culture medium every 2 days until retraction assay.
- Pour 1 ml of culture medium into each well (6-well plate).
- In vitro retraction assay
- Mix 10 μg of ephrinA3-Fc/Fc and 45 μg of anti-human Fc antibody in a microcentrifuge tube.
- Place the tube into a 25 °C incubator.
- After 1 h, place the pre-clustered solution into organotypic hippocampal slice culture medium. The final concentration for ephrinA3-Fc/Fc is 10 μg/ml.
- Place the plate into a 5% CO2 incubator at 37 °C.
- After 16 h, fix the hippocampal slices on the confetti discs with 4% PFA in PBS for 30 min at room temperature.
- Mount hippocampal slices onto a glass slide with Vectashield mounting media, and cover with a cover glass.
- Mix 10 μg of ephrinA3-Fc/Fc and 45 μg of anti-human Fc antibody in a microcentrifuge tube.
- Imaging and analysis
- Image dendritic spines in primary/secondary dendrites of hippocampal pyramidal neurons in CA1 region using a confocal microscope. Sequential z-images consisted of optical section (1,024 x 1,024 pixels) with 0.1 μm intervals using 63x oil immersion objective (numerical aperture, 1.3) with 9x digital zoom. Figure 4 shows z-stacked images of typical dendritic spines.
- Reconstruct z-images, and measure spine density, spine length, volume by the Imaris Filament Tracer.
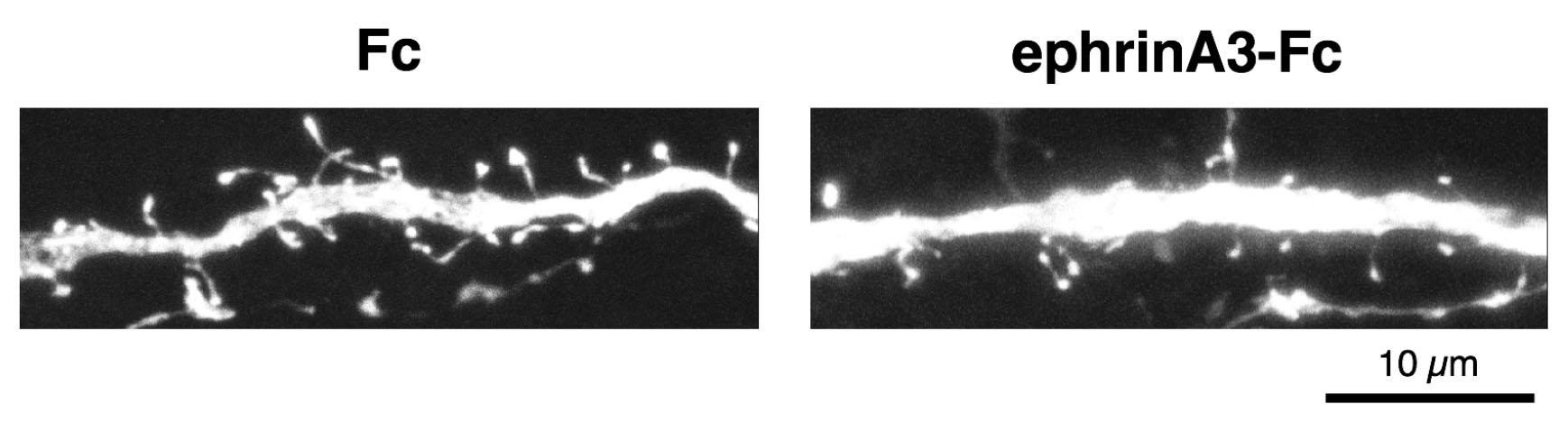 Figure 4. Example of results of dendritic spine retraction assay. Segments of dendrites from hippocampal slices, after treatment with ephrinA3-Fc (right) or Fc (left). Scale bar: 10 μm.
Figure 4. Example of results of dendritic spine retraction assay. Segments of dendrites from hippocampal slices, after treatment with ephrinA3-Fc (right) or Fc (left). Scale bar: 10 μm.
- Image dendritic spines in primary/secondary dendrites of hippocampal pyramidal neurons in CA1 region using a confocal microscope. Sequential z-images consisted of optical section (1,024 x 1,024 pixels) with 0.1 μm intervals using 63x oil immersion objective (numerical aperture, 1.3) with 9x digital zoom. Figure 4 shows z-stacked images of typical dendritic spines.
Recipes
- Culture medium
250 ml MEM with glutamax-1
125 ml horse serum
120 ml EBSS with 3% D-glucose
5 ml penicillin-streptomycin
0.3 ml nystatin - Slicing buffer
487.5 ml EBSS
12.5 ml 1 M HEPES
Acknowledgments
This protocol was adopted from Iwata et al. (2015). This work was supported by the JSPS KAKENHI (15H04263, 16K14559) and MEXT KAKENHI (15H01454).
References
- Iwata, R., Matsukawa, H., Yasuda, K., Mizuno, H., Itohara, S. and Iwasato, T. (2015). Developmental RacGAP α2-chimaerin signaling is a determinant of the morphological features of dendritic spines in adulthood. J Neurosci 35(40): 13728-13744.
- Mizuno, H., Luo, W., Tarusawa, E., Saito, Y. M., Sato, T., Yoshimura, Y., Itohara, S. and Iwasato, T. (2014). NMDAR-regulated dynamics of layer 4 neuronal dendrites during thalamocortical reorganization in neonates. Neuron 82(2): 365-379.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Iwata, R. and Iwasato, T. (2016). In vitro Assay for Dendritic Spine Retraction of Hippocampal Neurons with Sparse Labeling. Bio-protocol 6(18): e1937. DOI: 10.21769/BioProtoc.1937.
-
Iwata, R., Matsukawa, H., Yasuda, K., Mizuno, H., Itohara, S. and Iwasato, T. (2015). Developmental RacGAP α2-chimaerin signaling is a determinant of the morphological features of dendritic spines in adulthood. J Neurosci 35(40): 13728-13744.
Category
Neuroscience > Cellular mechanisms
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link