- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protein Expression, Purification and Crystallization of the Sxl-Unr-msl2 Ribonucleoprotein Complex
Published: Vol 6, Iss 17, Sep 5, 2016 DOI: 10.21769/BioProtoc.1917 Views: 10245
Reviewed by: Arsalan DaudiLongping Victor TseAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Identification of Intrinsic RNA Binding Specificity of Purified Proteins by in vitro RNA Immunoprecipitation (vitRIP)
Marisa Müller [...] Peter B. Becker
Mar 5, 2021 4649 Views
Immunoprecipitation for Protein-Protein Interactions and for RNA Enrichment in Drosophila melanogaster
Giulia Romano [...] Fabian Feiguin
Dec 5, 2021 3364 Views

A Step-by-Step Computational Protocol for Functional Annotation and Structural Modelling of Insect Chemosensory Proteins
Rajeswari Kalepu [...] Nor Azlan Nor Muhammad
Nov 20, 2025 1667 Views
Abstract
This protocol describes the expression, purification and crystallization of a ternary protein-protein-RNA complex, consisting of the two RNA recognition motifs (RRMs) of Sex-lethal (Sxl), the first of five cold shock domains of Upstream-of-N-Ras (Unr), and an 18-nucleotide region of msl2 mRNA, called the F fragment (Hennig et al., 2014).The biological role of the complex is the translational repression of msl2 mRNA, preventing the formation of the dosage compensation complex and subsequent 2-fold hypertranscription of X-linked genes in Drosophila females. As orthologous RRM-containing proteins and Unr exist in humans, similar complexes potentially also form during translational repression in vertebrates. The protocol describes the in vitro assembly of the complex and its purification followed by crystallization for X-ray crystallography structure determination. Part of the protocol has been published elsewhere (Hennig et al., 2013 and 2014), but some parts are described here in more detail.
Materials and Reagents
- Membrane dialysis tubing Spectra/Por 6 prewetted, 1.1 ml/cm 3,500 Da cut-off (Spectrum, Spectra/Por®, catalog number: 132590 )
- Membrane dialysis tubing Spectra/Por 6 prewetted, 1.1 ml/cm 10,000 Da cut-off (Spectrum, Spectra/Por®, catalog number: 128118 )
- Amicon® Ultra-15 centrifugal filter unit with Ultracel-3 membrane (Merck Millpore, catalog number: UFC900308 )
- Amicon® Ultra-15 centrifugal filter unit with Ultracel-10 membrane (Merck Millpore, catalog number: UFC901008 )
- Amicon® Ultra-4 centrifugal filter unit with Ultracel-3 membrane (Merck Millpore, catalog number: UFC800324 )
- Amicon® Ultra-4 centrifugal filter unit with Ultracel-10 membrane (Merck Millpore, catalog number: UFC801024 )
- EasyXtal 15-well tool (Qiagen, catalog number: 132006 )
- 0.22 µm filter
- 96-well plate
- SS34 centrifugation tubes
- Escherichia coli (E. coli) Bl21 (DE3)from stocks (stored at -80 °C)
- pTRX-Sxl and pTRX-CSD plasmids (available upon request)
- Tobacco etch virus (TEV) protease (1 mg/ml stock solution, in-house production)
- Precision plus protein all blue prestained protein standard (Bio-Rad Laboratories, catalog number: 1610373 )
- LB medium
- Kanamycin (33 mg/ml stock solution, stored at -20 °C)
- Isopropyl-β-D-1-thiogalactopyranoside (IPTG) (1 M stock solution)
- Ni-NTA resin (Qiagen, catalog number: 30230 )
- Sodium phosphate (molecular biology grade)
- Sodium chloride (molecular biology grade)
- Potassium phosphate (molecular biology grade)
- Imidazole (molecular biology grade, > 99%)
- β-mercaptoethanol
- Sodium azide
- Ammonium sulfate (molecular biology grade, > 99% purity)
- 18-mer RNA based on E fragment of msl2 mRNA: UUU UUU UGA GCA CGU GAA (synthesized) (IBA) (UniProt Consortium, catalog number: P50534 )
- Lithium sulfate (X-ray crystallography grade, > 99% purity)
- PEG 3350 (Sigma-Aldrich, catalog number: 39713 )
- Dithiothreitol (DTT) (> 99% purity)
- Glycerol (Sigma-Aldrich, catalog number: 49767 )
- Lysis buffer (see Recipes)
- Wash buffer A (see Recipes)
- Wash buffer B (see Recipes)
- Elution buffer (see Recipes)
- NMR buffer (see Recipes)
- Running buffer (see Recipes)
Equipment
- Erlenmeyer flasks (500 ml)
- FPLC protein purifier
- Superdex S75 gel filtration size-exclusion chromatography column HiLoad 26/600 (GE Healthcare)
- Nanodrop system
Procedure
- Expression of Sxl and Unr
- The pTRX-Sxland and pTRX-CSD plasmids [(based on the pETM11 expression vector) including the nucleotide sequence of RRM1-RRM2 of Sxl, corresponding to residues 123-294 of D. melanogaster Sxl, UniProt entry: P19339, and the first cold shock domain of Unr (CSD1), corresponding to residues 181-252 of D. melanogaster Unr, UniProt entry: Q9VSK3, respectively. Both plasmids have an N-terminal His6-tagged thioredoxin solubility tag, connected to Sxlor Unrwith a TEV-cleavage site (see Figure 1)] were transformed into the Escherichia coli strain Bl21 (DE3), using heat shock for 2 min at 42 °C. The expression is efficient and likely other expression strains will also work. Also, the expression of both constructs is highly reproducible. Each time more proteins were needed a fresh transformation has been performed.
- For large-scale protein production one overnight culture of 50 ml LB medium with 33 µg/ml Kanamycin was inoculated with a single colony of each transformation plate (Sxl and Unr). The cultures were incubated at 37 °C and 220 rpm for around 16 h.
- Afterwards, 1 L LB medium with 33 µg/ml Kanamycin, divided into two 2 L Erlenmeyer flasks (500 ml each), was inoculated with 2 ml of the overnight culture (ratio 1:250) and further incubated at 37 °C and 220 rpm.
- Upon reaching an optical density (OD600nm) between 0.8-1 (after approximately 3-4 h), the protein production was induced by addition of IPTG to a final concentration of 1 mM. After induction, the cultures were incubated at 20 °C and 220 rpm for 16 h.
- The cells were then harvested at 3,000 x g for 30 min at 4 °C.
- The cell pellets were resuspended in 45 ml lysis buffer and transferred to a 50 ml tube before sonication for 5 min (1 sec intervals, microtip) on ice.
- Cell debris was pelleted by centrifugation for 30 min at 5,000 x g and 4 °C.
- The supernatant, containing overexpressed Sxl or Unr was filtered through a 0.22 µm filter before purification.
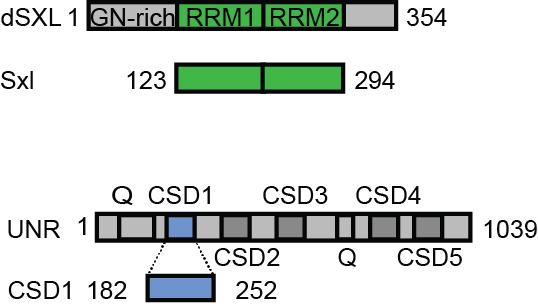
Figure 1. Sxl and Unr constructs used in this protocol. Amino acid numbers according to their native sequence are indicated (Q, glutamine-rich domain).
- The pTRX-Sxland and pTRX-CSD plasmids [(based on the pETM11 expression vector) including the nucleotide sequence of RRM1-RRM2 of Sxl, corresponding to residues 123-294 of D. melanogaster Sxl, UniProt entry: P19339, and the first cold shock domain of Unr (CSD1), corresponding to residues 181-252 of D. melanogaster Unr, UniProt entry: Q9VSK3, respectively. Both plasmids have an N-terminal His6-tagged thioredoxin solubility tag, connected to Sxlor Unrwith a TEV-cleavage site (see Figure 1)] were transformed into the Escherichia coli strain Bl21 (DE3), using heat shock for 2 min at 42 °C. The expression is efficient and likely other expression strains will also work. Also, the expression of both constructs is highly reproducible. Each time more proteins were needed a fresh transformation has been performed.
- Purification of Sxl and Unr-CSD1
- Purification of Sxl
- A Ni-NTA gravity flow column, filled with 2 ml of Ni-NTA resin was equilibrated in lysis buffer with 10 column volumes (CVs).
- After addition of the supernatant, the column was washed with 10 CVs of lysis buffer, 10 CVs of wash buffer A, and 10 CVs of wash buffer B.
- Sxl was eluted with 5 CVs of elution buffer in 2 ml fractions, and most of the protein is in the first two fractions (see Figure 2A).
- The His6-thioredoxin-tag was then cleaved off with 1 mg TEV protease and 10 mM β-mercaptoethanol during dialysis of the entire elution volume (10 ml with around 3 mg/ml protein concentration) against 4 L of wash buffer B overnight, using a 1.1 ml/cm dialysis tube with a 10 kDa cut-off (removing excess imidazole and β-mercaptoethanol).
- The tag was then separated from Sxl by using a second Ni-NTA column equilibrated with 10 CVs in wash buffer B. The flow-through, containing Sxl was collected and applied a second time on the Ni-NTA column, where again the flow-through was collected (see Figure 2B).
- The flow-through was then dialyzed overnight against NMR buffer in a 1.1 ml/cm dialysis tube with 10 kDa cut-off.
- Sxl was further purified, using size-exclusion chromatography on a SuperdexS75 HiLoad 26/600 gel filtration column equilibrated with 4 CVs in running buffer. The flow rate of running buffer during purification was 0.5 ml/min. Three wavelengths (190 nm, 215 nm, and 280 nm) were used to monitor the protein elution profile.
- Sxl containing fractions (2 ml fractions were collected in an automated fashion in 96-well plate format) were pooled and 10 mM DTT, 0.02% sodium azide were added. The usual yield was around 30 mg per liter expression medium.
- Sxl was then used to prepare the ternary complex (see section C).
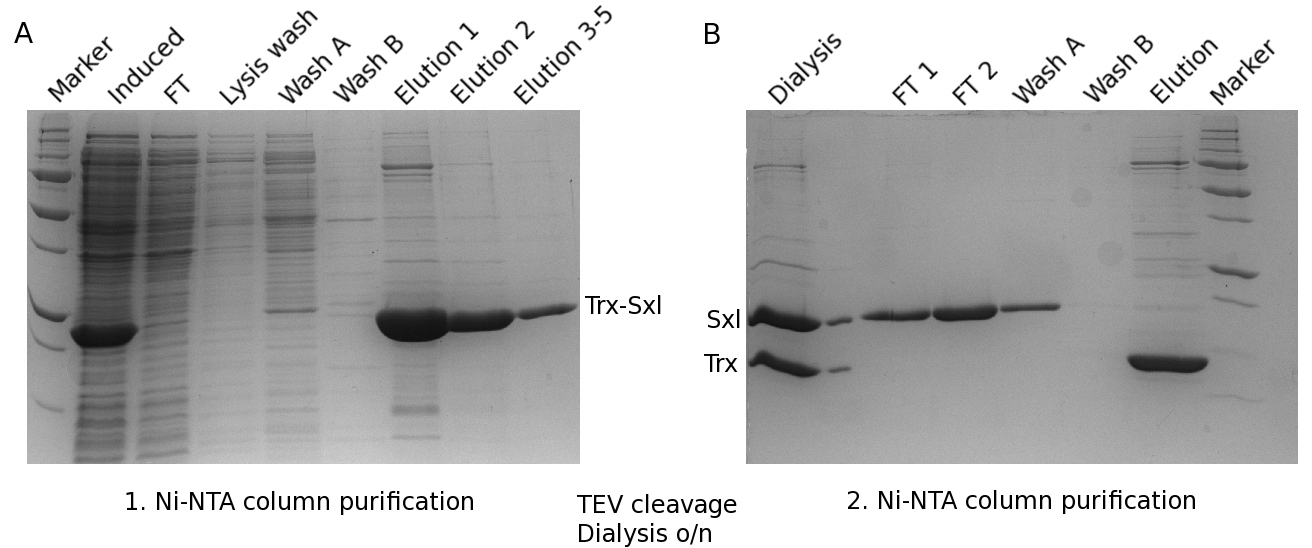
Figure 2. SDS-PAGE monitoring the Sxl purification process. A. SDS-PAGE gel (according to Laemmli, see Reference 3) of the first Ni-NTA column purification of Sxl. FT means flow-through. Each elution fraction (elution 1 to 5 in gel legend) was 2 ml. B. SDS-PAGE gel of the second Ni-NTA column purification of Sxl using the same marker.
- A Ni-NTA gravity flow column, filled with 2 ml of Ni-NTA resin was equilibrated in lysis buffer with 10 column volumes (CVs).
- Purification of Unr-CSD1
- See steps a-e in section B1. These steps are identical except that a dialysis tube with a 3.5 kDa cut-off is used for Unr-CSD1 (see Figure 3).
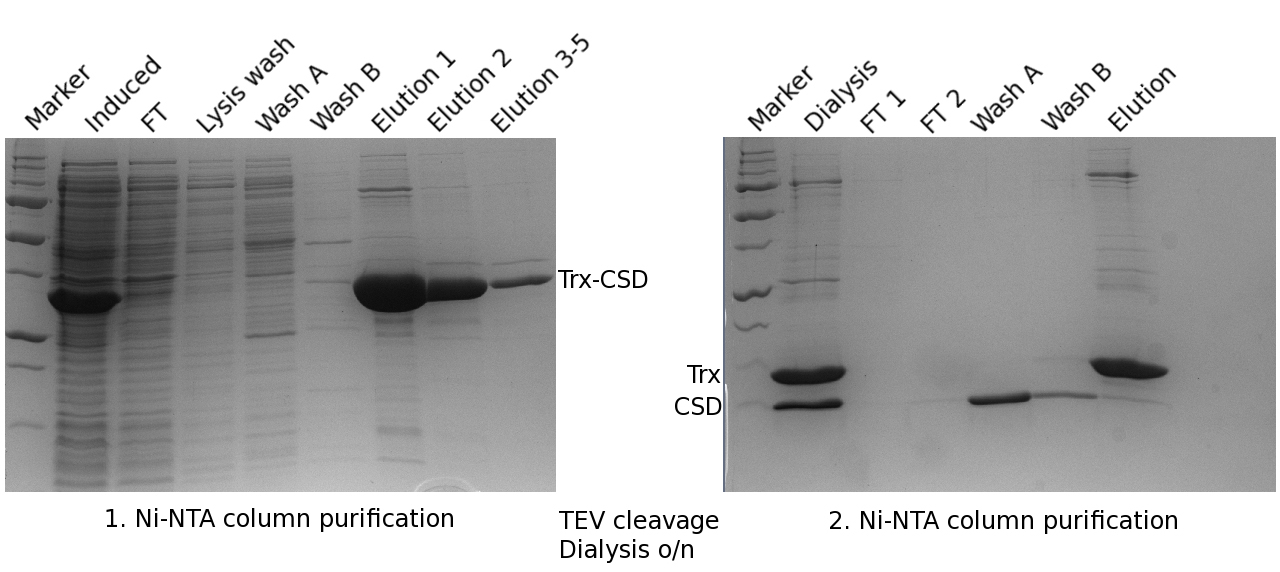
Figure 3. SDS-PAGE gel (according to Laemmli, see Reference 3) of the first (left) and second (right) Ni-NTA column purification of Unr-CSD. Each elution fraction (elution 1 to 5 in gel legend) was 2 ml. The same marker as in Figure 2 was used. FT means flow-through. - Unr-CSD1 binds strongly to bacterial nucleic acids, which need to be separated. To do this, ammonium sulfate precipitation was used. The 10 ml flow through from the second Ni-NTA, containing Unr-CSD1 was filled into a 100 ml beaker with a magnetic stirrer. The volume was increased to 20 ml with wash buffer B.
- While stirring (around 150 rpm), ammonium sulfate was added. In a first step directly 70 g were added. The solubility of ammonium sulfate is 74.4 g/100 ml at 20 °C. After addition and dissolving of ammonium sulfate the volume increases almost to 100 ml (important to consider when choosing the beaker size).
- More ammonium sulfate was added step-wise(the amount per step is not critical but large excess above the solubility limit given above should be avoided to prevent longer and/or repeated dialysis and/or larger dialysis volume), until the solution got milky and ammonium sulfate was not further dissolvable, indicative of saturation.
- The solution was filled into SS34 centrifugation tubes and centrifuged at 20,000 x g for 20 min at 4 °C. The supernatant was discarded and the pellet was resuspended in 10 ml of NMR buffer (the ammonium sulfate precipitation is also useful for changing the pH from pH 8 during Ni-NTA purification via pI 7.8 to the pH 6 of the NMR buffer).
- Unr-CSD1 was now dialyzed overnight against 4 L NMR buffer in a 1.1 ml/cm dialysis tube with a 3.5 kDa cut-off.
- Followed by further purification, using size-exclusion chromatography on a SuperdexS75 HiLoad 26/600 gel filtration column equilibrated with 4 CVs in running buffer. The flow rate of running buffer during purification was 0.5 ml/min. Three wavelengths (190 nm, 215 nm, and 280 nm) were used to monitor the protein elution profile.
- Unr-CSD1 containing fractions (2 ml fractions were collected in an automated fashion in 96-well plate format) were pooled and 10 mM DTT, 0.02% sodium azide were added. The usual yield was around 15 mg per liter expression medium.
- Unr-CSD1 was then used to prepare the ternary complex (see section C).
- See steps a-e in section B1. These steps are identical except that a dialysis tube with a 3.5 kDa cut-off is used for Unr-CSD1 (see Figure 3).
- Purification of Sxl
- Assembly and purification of the ternary Sxl-Unr-msl2 mRNA complex
- Both, Sxl and Unr-CSD1 solutions were diluted with NMR buffer to around 20 µM and mixed to an equimolar ratio (at higher concentrations > 100 µM precipitation has been observed).
- The 18-mer RNA (see Materials and Reagents) was then added in a 1:1.1 excess (protein: RNA). The order of complex assembly is important as Sxl without Unr-CSD1 tends to precipitate upon addition of RNA longer than 10 nucleotides.
- The complex solution was then concentrated down to a volume of 3 ml using a 3 kDa cut-off concentrator.
- Size-exclusion chromatography was used using a SuperdexS75 HiLoad 26/600 gel filtration column in running buffer to remove possible excess of single components. Right-angle light scattering was used to assess the molecular weight of the complex (see Figure 4). All steps can be performed at room temperature except centrifugation which should be done at 4 °C, to avoid heating and precipitation of the sample.
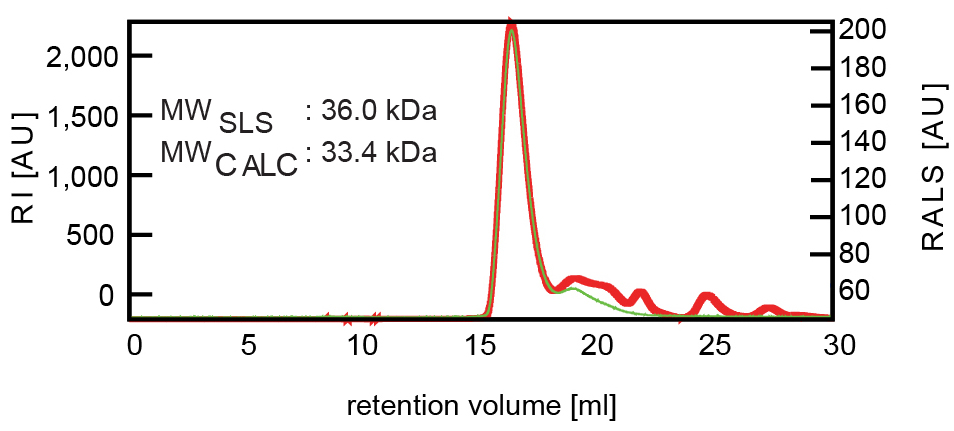
Figure 4. FPLC-RALS chromatogram of the Sxl-Unr-CSD-msl2 mRNA ternary complex. The refractive index (RI) is shown as a red line, and the right-angle light scattering value (RALS) is shown as a green line. From this data, the molecular weight of the complex was calculated to 36 kDa. - Fractions containing the complex were pooled and 10 mM DTT were added.
- Pooled fractions were concentrated using a 3 kDa cut-off concentrator to a concentration of 15 mg/ml (450 µM).
- The concentration determination was done using absorbance at 280 nm wavelength on a Nanodrop system and calculated extinction coefficients. The lack of tryptophans for Unr-CSD1 made the concentration determination less accurate.
- All gel filtration and centrifugation steps were done at 4 °C. All other steps, including Ni-NTA purification and mixing of components, were performed at room temperature. Intermediate products can be stored at 4 °C. We recommend avoiding freezing as we observed slight changes of isothermal titration calorimetry curves after thawing, for which we do not have an explanation. Although both proteins are considered to be stable longer breaks than overnight during sample preparation should be avoided.
- Both, Sxl and Unr-CSD1 solutions were diluted with NMR buffer to around 20 µM and mixed to an equimolar ratio (at higher concentrations > 100 µM precipitation has been observed).
- Crystallization of the ternary Sxl-Unr-msl2 mRNA complex
- Hanging drop crystallization was used to crystallize the complex. Drops were setup by pipetting 2 µl of the complex solution from step C6 onto the inside of the lid of the crystallization well, followed by carefully pipetting 2 µl of the reservoir solution onto the first drop, without mixing!
- The reservoir solution was optimized to be 0.1 M LiSO4 and 1% PEG 3350 (w/v), of which 500 µl were pipetted into the well.
- The plate is then stored at 4 °C.
- Tiny crystals only appear after two months and they need another month to grow to a final size shown in Figure 5. It is important to note, that the slow growth is likely due to the presence of DTT, as an inter-complex disulphide bridge is formed between cysteines of Unr-CSD1, which is important for crystallization. The disulphide bridge is formed slowly due to slower oxidation of DTT at 4 °C and the sealed hanging drop well. This is speculative, but crystals did not form when no DTT was used.
- As cryoprotectant for storage 30% glycerol was used.
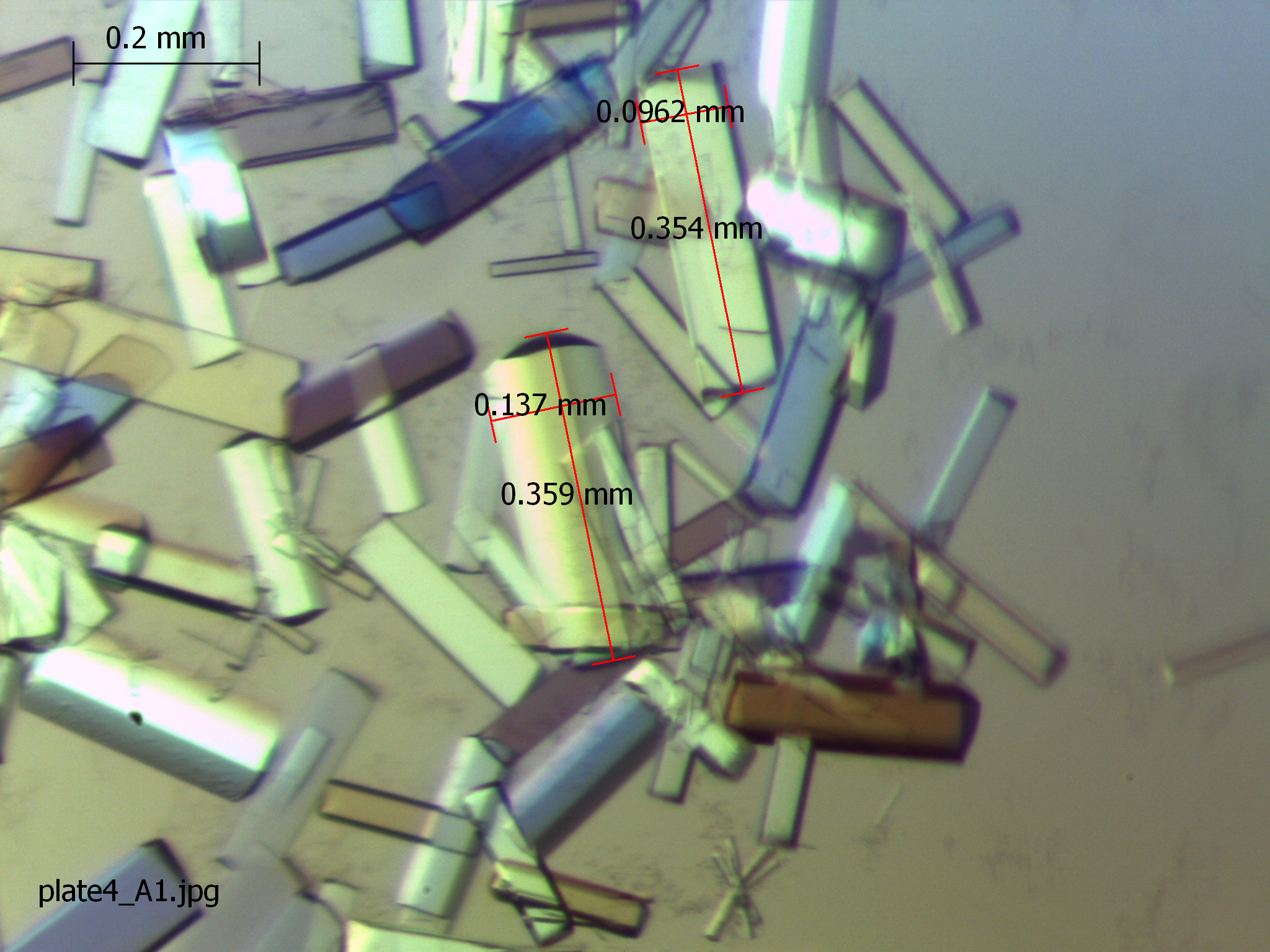
Figure 5. Crystals obtained from crystallization conditions described in this protocol. The size of two crystals is indicated in the picture as well as a scale bar.
- Hanging drop crystallization was used to crystallize the complex. Drops were setup by pipetting 2 µl of the complex solution from step C6 onto the inside of the lid of the crystallization well, followed by carefully pipetting 2 µl of the reservoir solution onto the first drop, without mixing!
Recipes
- Lysis buffer
50 mM sodium phosphate
300 mM NaCl
pH 8.0 - Wash buffer A
50 mM sodium phosphate
300 mM NaCl
10 mM imidazole
pH 8.0 - Wash buffer B
50 mM sodium phosphate
300 mM NaCl
20 mM imidazole
pH 8.0 - Elution buffer
50 mM sodium phosphate
300 mM NaCl
120 mM imidazole
pH 8.0 - NMR buffer
10 mM potassium phosphate
50 mM NaCl
10 mM DTT
pH 6.0 - Running buffer
10 mM potassium phosphate
50 mM NaCl
pH 6.0
Acknowledgments
We thank the crystallization facility at the Max-Planck-Institute for Biochemistry, Martinsried for initial crystallization screens and Arie Geerlof for protein expressions and purifications. J.H. acknowledges postdoctoral fellowships from the Swedish Research Council (Vetenskapsradet) and the European Molecular Biology Organization (EMBO, ALTF276-2010). M.S. acknowledges the Deutsche Forschungsgemeinschaft (DFG), grants SFB1035 and GRK1721.
References
- Hennig, J., Wang, I., Sonntag, M., Gabel, F. and Sattler, M. (2013). Combining NMR and small angle X-ray and neutron scattering in the structural analysis of a ternary protein-RNA complex. J Biomol NMR 56(1): 17-30.
- Hennig, J., Militti, C., Popowicz, G. M., Wang, I., Sonntag, M., Geerlof, A., Gabel, F., Gebauer, F. and Sattler, M. (2014). Structural basis for the assembly of the Sxl-Unr translation regulatory complex. Nature 515(7526): 287-290.
- Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227(5259): 680-685.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hennig, J. and Sattler, M. (2016). Protein Expression, Purification and Crystallization of the Sxl-Unr-msl2 Ribonucleoprotein Complex. Bio-protocol 6(17): e1917. DOI: 10.21769/BioProtoc.1917.
Category
Biochemistry > Protein > Structure
Biochemistry > RNA > RNA-protein interaction
Biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










