- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Single-cell Visualization of Chromosome Transcriptional Territories by RNA-paint
Published: Vol 6, Iss 17, Sep 5, 2016 DOI: 10.21769/BioProtoc.1914 Views: 9398
Reviewed by: Anonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Temporally and Spatially Controlled Age-Related Prostate Cancer Model in Mice
Sen Liu [...] Qiuyang Zhang
Jan 5, 2025 2702 Views

Fast TV-PRO-seq: Accelerated and Streamlined Protocol for Timing RNA Polymerase Pausing
Jie Zhang [...] Shaohui Zhang
Jul 20, 2025 2425 Views

Efficient Fluorescent Labeling of Human Trophoblast Stem Cells via a CRISPR/Cas9-Mediated Knock-In Approach in a Safe Harbor Locus
Hengshan Zhang [...] Danny J. Schust
Jan 5, 2026 307 Views
Abstract
We developed a FISH-based method to directly assess chromosome-wide transcriptional activity, thereby enabling the visualization of the actively transcribed fraction of a chromosome at the single-cell level. We applied this method to probe the activity of X-chromosomes and its instability in the context of human embryonic stem cells and cancer cells.
Keywords: TranscriptionMaterials and Reagents
- 24-well plates (Sigma-Aldrich, catalog number: Z707791-126EA )
- 13 mm round coverslips (Thermo Fisher Scientific, catalog number: 174950 )
- Glass slides
- Filter (0.2 μm)
- Filtration unit (Merck Millipore, catalog number: SCGPS05RE )
- Human embryonic stem cells (H9, WIBR2 and HUES1) and cancer cells (TCCSUP and RT112)
- Matrigel
- PBS solution (Life Technologies)
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- Sucrose (Sigma-Aldrich, catalog number: S0389 )
- Magnesium chloride (MgCl2) (Sigma-Aldrich, catalog number: M8266 )
- PIPES (Sigma-Aldrich, catalog number: P6757 )
- NaOH
- Triton X-100 (Sigma-Aldrich, catalog number: X100 )
- Ribonucleoside vanadyl complex (New England Biolabs, catalog number: S1402 )
- Paraformaldehyde (PFA) (16%, EM grade) (VWR International, catalog number: 100503-916 )
- EtOH
- Human Cot-1 DNA (Life Technologies)
- Sodium acetate anhydrous (NaOAc) (Sigma-Aldrich, catalog number: W302406 )
- Deionized formamide (Sigma-Aldrich, catalog number: F9037 )
- Rubber cement
- 20x SSC (Sigma-Aldrich, catalog number: 93017 )
- 20 mg/ml BSA (New England Biolabs, catalog number: B9000S )
- Dextran sulfate (Sigma-Aldrich, catalog number: 67578 )
- Formamide (Sigma-Aldrich, catalog number: 47670 )
- Mounting medium (Vector Laboratories, catalog number: H-1200 )
- Cy3-labelled human X-Chromosome paint (Metasystem, catalog number: D-0323-050-OR )
- FITC-labelled human X-Chromosome paint (Metasystem, catalog number: D-0323-050-FI )
- CSK buffer (see Recipes)
- 3% PFA/PBS solution (see Recipes)
- Hybridisation buffer (2x) (see Recipes)
- Washing solution (50% formamide/2x SSC) (see Recipes)
- Denaturating solution (70% formamide/2x SSC) (see Recipes)
Equipment
- Hybridisation table (Boekel Scientific, model: 240000 )
- Shake 'N BakeTM hybridisation oven (Boekel Scientific, model: 136400 )
- Centrifuge (VWR, Ependorf®, model: 5417R )
- Millivac-mini vacuum pump (Merck Millipore, catalog number: XF5423050 )
- Ependorf® Thermomixer® R (Sigma-Aldrich, catalog number: T3317 )
Note: This product has been discontinued. - Fluorescent microscope (motorized stage) (Leica, model: DMI-6000 )
Procedure
- Prepare 24-well plates with 13 mm round coverslips at the bottom of the wells. Coat coverslips with matrigel for human embryonic stem cells. Split cells in colonies with a ratio of 1:5 on coverslips. Grow cells for 24 to 48 h.
- Wash cells with 1 ml PBS per well.
- Incubate cells for 5 min on ice with 0.5 ml per well of ice-cold CSK buffer freshly supplemented with 0.5% Triton X-100 and 2 mM vanadyl ribonucleoside complex. The core CSK buffer can be kept at 4 °C several months.
- Fix cells for 10 min at room temperature with 0.5 ml per well of PBS/3% PFA supplemented with 2 mM vanadyl ribonucleoside complex.
- Rinse cells with 1 ml per well 3 times with cold EtOH (70%) for 4 min.
- Either proceed to step 8 or the protocol can be stopped here by tightly wrapping the 24-well plate with parafilm, stretching it gently to ensure good fit and avoid evaporation. Cells can be stored at -20 °C for one month.
- On the day of the experiment, bring coverslips to 4 °C in 70% EtOH.
- For probe preparation, for each coverslip, add 290 µl of water to 5 µl of concentrated chromosome paint and 5 µl of human Cot-1 DNA (1 µl/µg). Precipitate DNA by adding 1/10 3 M NaOAc and 2.5 volumes of 100% EtOH and storing tubes overnight at -20 °C. Probes are prepared individually for each coverslip: 1 mix for 1 coverslip.
- Spin probes at 15,000 x g at 4 °C for 20 min. The pellet should be visible and is washed 2 times with 70% EtOH and spun for 10 min at 15,000 x g at 4 °C.
- Resuspend the pellet in 5 µl of 50% deionized formamide/50% hybridisation buffer (2x), and incubate for 10 min at 37 °C.
- Denature the chromosome paint 7 min at 75 °C, incubate for 30 min at 37 °C and store it on ice.
- In the meantime, dehydrate coverslips in 90% and 100% EtOH for 4 min each. Coverslips are then air dried on Kimwipes, cells face up. Heat clean glass slides on the hybridisation table at 37 °C, where the moats have been prefilled with water.
- Dispense 5 µl of probe per coverslip on glass slides, and place coverslips on the probe, with cells facing down (Figure 1). Incubate the coverslips at 37 °C overnight, and evaporation around the coverslips is prevented using rubber cement to seal the coverslips to the glass slides.
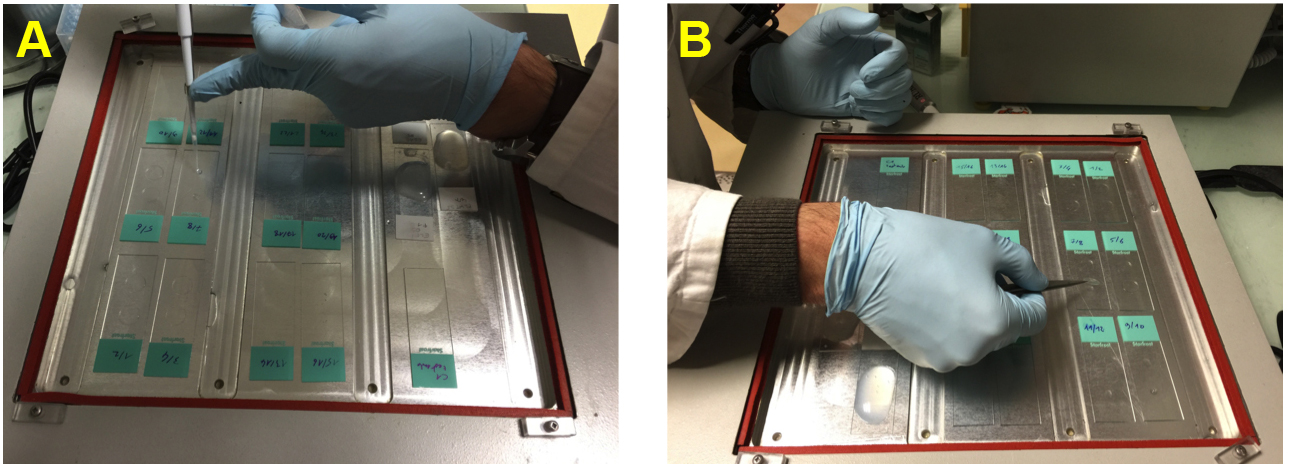
Figure 1. Manipulating coverslips. A. Putting 5 µl of probe on slides. B. Putting coverslips with cells face down on probe preparation. - The next day, remove rubber cement by gently drawing it from the coverslips. Spill 2 ml of 2x SSC over the coverslips and glass slides, and gently start pushing around the coverslips by making them float in 2x SSC. Fully detached coverslips are transferred to 24-well plates for further washes. Wash coverslips 3 times with 1 ml per well of 50% formamide/2x SSC, and 3 times in 2x SSC, all washes at 42 °C. Incubate 24-well plates with coverslips in hybridization oven with low agitation (shaker speed set at medium speed).
- Mount coverslips on glass sides using 5 µl of mounting medium containing DAPI. Use transparent nail polish to seal the coverslips to the slides.
- Observe fluorescent signals with a fluorescent microscope (magnification 100x).
- For dual RNA-FISH involving chromosome paint (Vallot et al., 2015)
- RNA-FISH is performed in two-steps. First, RNA-FISH with chromosome paint is performed overnight as described above (steps 1-14).
- Coverslips are washed 3-times in 50% formamide/2x SSC at 37 °C and re-incubated overnight with the second probe.
- Coverslips are washed 3-times in 50% formamide/2x SSC and 3-times in 2x SSC at 37 °C. Observe both signals simultaneously.
- RNA-FISH is performed in two-steps. First, RNA-FISH with chromosome paint is performed overnight as described above (steps 1-14).
- For successive RNA/DNA-FISH with chromosome paint (Vallot et al., 2015)
- RNA-FISH is performed on slides as described above using a Cy3-labelled paint to visualize RNA, take pictures and note cell coordinates using a stage-motorized Leica microscope.
- Wash slides, cells are permeabilized for 10 min in 0.1 N HCl/0.7% Triton X-100 on ice, treated with 100 µg/µl RNase in 2x SSC for 1 h at 37 °C.
- Denature slides for 10 min at 80 °C in 70% formamide/2x SSC.
- DNA is visualized using a FITC-labelled X chromosome paint, it is denatured for 2 min at 75 °C and incubated with slides overnight.
- Wash slides 3 times with 2x SSC at 45 °C and 0.1x SSC at 60 °C.
- For dual RNA-FISH involving chromosome paint (Vallot et al., 2015)
Representative data
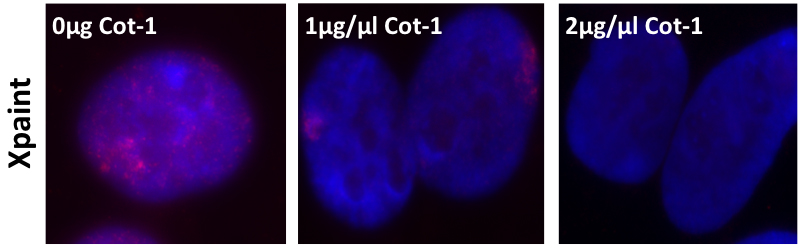
Figure 2. Optimizing the X-chromosome paint RNA FISH. 5 µl of cy3-X-chromosome paint are co-precipitated with various amounts of human Cot-1 DNA prior to over-night hybridisation. Optimization of the quantity of Cot-1 should be performed when changing supplier for chromosome paint.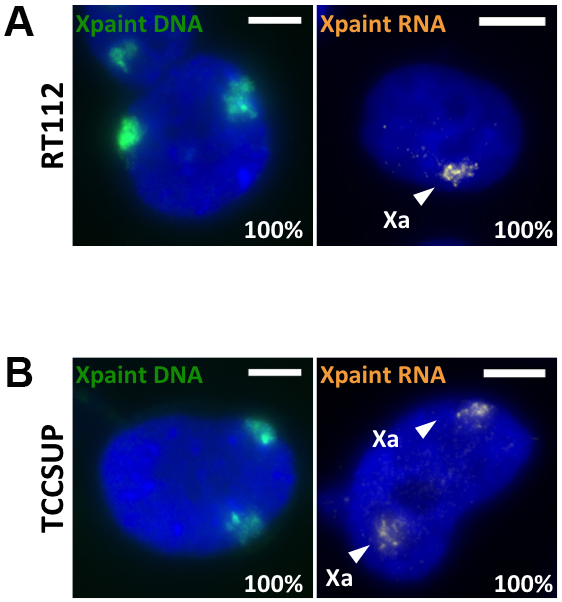
Figure 3. Single-cell detection of X-chromosome activity in cancer cells. X-chromosome content of two bladder cancer cell lines RT112 (A) and TCCSUP (B) was assessed by DNA-FISH using FITC-labeled X-chromosome paint (left panel) and X-chromosome activity by RNA-paint using cy3-labeled X-chromosome paint (right panel).The percentage in the right lower corner indicates the percentage of nuclei with displayed pattern.
Recipes
- CSK buffer
100 mM NaCl
300 mM sucrose
3 mM MgCl2
10 mM PIPES
Adjust pH to 6.8 with NaOH to facilitate dissolution of chemicals in water
Filter (0.2 μm) - 3% PFA/PBS solution
9.4 ml of 16% PFA solution
40.6 ml of PBS - Hybridisation buffer (2x)
4x SSC
4 mg/ml BSA
20% dextran sulfate
40 mM vanadyl ribonuclside complex - Washing solution (50% formamide/2x SSC) (50 ml), pH = 7.2
25 ml of formamide
5 ml of 20x SSC
20 ml of H2O
60 µl of 2.5 N HCl - Denaturating solution (70% formamide/2x SSC) (50 ml), pH = 7.2
35 ml of formamide
5 ml of 20x SSC
10 ml of H2O
145 µl of 2.5 N HCl
Acknowledgments
We thank members of our laboratory for stimulating discussion. The research leading to these results has received funding from the European Research Council under the European Community’s Seventh Framework Programme (FP7/2007-2013)/ERC grant agreement No. 206875 and from the Inserm (Avenir Program R0721HS).
References
- Vallot, C., Ouimette, J. F., Makhlouf, M., Feraud, O., Pontis, J., Come, J., Martinat, C., Bennaceur-Griscelli, A., Lalande, M. and Rougeulle, C. (2015). Erosion of X chromosome inactivation in human pluripotent cells initiates with XACT coating and depends on a specific heterochromatin landscape. Cell Stem Cell 16(5): 533-546.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Vallot, C. and Rougeulle, C. (2016). Single-cell Visualization of Chromosome Transcriptional Territories by RNA-paint. Bio-protocol 6(17): e1914. DOI: 10.21769/BioProtoc.1914.
Category
Stem Cell > Embryonic stem cell > Cell-based analysis
Molecular Biology > DNA > Gene expression
Molecular Biology > RNA > Transcription
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link








