- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Chromium-51 (51Cr) Release Assay to Assess Human T Cells for Functional Avidity and Tumor Cell Recognition
Published: Vol 6, Iss 16, Aug 20, 2016 DOI: 10.21769/BioProtoc.1906 Views: 19802
Reviewed by: Masahiro MoritaMichael EnosEmilie Besnard

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

In vivo OVA-specific Cytotoxic CD8+ T Cell Killing Assay
Nada Chaoul [...] Claude Leclerc
Jun 20, 2016 24220 Views

Measuring Procaspase-8 and -10 Processing upon Apoptosis Induction
Sabine Pietkiewicz [...] Inna N. Lavrik
Jan 5, 2017 10552 Views

Monitoring Natural Killer Cell Function in Human Ovarian Cancer Cells of Ascitic Fluid
Vikash Kumar and Asok Mukhopadhyay
Dec 20, 2018 6932 Views
Abstract
Cytotoxic CD8+ T cells are able to specifically recognize and kill target cells through specific interaction between their T cell receptors (TCRs) and small immunogenic peptides (antigens) presented by major histocompatibility complex (MHC) molecules. The antigen recognition capacity and in vitro lytic activity of antigen-specific cytotoxic T cells can be assessed functionally in the so-called chromium 51 (51Cr) release assay, which was developed almost 50 years ago in our institution (Brunner et al., 1968). Radioactively-labelled cells deficient for endogenous antigen presentation [e.g., transporter for antigen presentation (TAP)-deficient T2 cells] and stably transfected with the MHC of interest (e.g., HLA-A2+) are typically used as targets during this 4h assay. Alternatively, 51Cr-labelled virus-infected or tumor cell lines presenting immunogenic antigens endogenously can serve as target cells (e.g., for the assessment of tumor recognition).
In a peptide titration assay (section A), radioactively labelled target cells are pulsed with a serial dilution of the antigenic peptide and incubated at an effector (e.g., a CD8+ T cell clone) to target (51Cr -T2 cells) ratio (E:T) of 10:1 in a 96-well V-bottom plate for 4 h at 37 °C. In a tumor killing assay (section B), cytotoxic CD8+ effector cells are incubated at different ratios with the 51Cr-labelled target cell line (typically at E:T ratios of 30:1, 10:1, 3:1 and 1:1) in the presence or absence of the specific antigenic peptide (1 μM) and incubated for 4 h at 37 °C. At the end of the test, the amount of radioactivity release from the lysed target cells is determined in the supernatant using a liquid scintillation counter. The percentage of specific lysis, as well as the EC50 (i.e., 50% of maximal killing) and EMax values are then calculated, providing quantitative information about the antigen-specific functional avidity (i.e., the relative efficiency of T cell function based on antigen recognition via a defined TCR and maximal killing capacity of the analyzed T cells).
Materials and Reagents
- 96-well plates, V-shape (Corning, Costar®, catalog number: 3894 )
- 15 ml Falcon tubes (e.g., Fisher Scientific, catalog number: 14-959-53A )
- LumaPlate-96 (PerkinElmer, catalog number: 6006633 )
- Bassin or reservoir (e.g., Vitaris, catalog number: 4304-INT or equivalent)
- Antigen presenting target cells: e.g., human T2 (CRL-1992), mice P815 (ATCC, catalog number: TIB-64 ) or a characterized tumor cell line [e.g., Me275 (Zippelius et al., 2002)]
- Effector cells [(i.e., polyclonal or clonal population of antigen-specific cytotoxic CD8 T cells directly isolated from the blood of patients or healthy donors (Speiser et al., 2008)]
- RPMI 1640 (Thermo Fisher Scientific, Gibco®, catalog number: 61870010 )
- Fetal bovine serum (FBS) inactivated at 56 °C for 1 h (Thermo Fisher Scientific, Gibco®, catalog number: 10270106 )
- Antigenic peptide solution [e.g., NY-ESO-1157-165 peptide (SLLMWITQC)] dissolved in 1x PBS at initial concentration 1 mg/ml
Note: For the controls, an irrelevant peptide solution at 1 mg/ml that is not recognized by the T cells (i.e., any other class I short peptide). - 51Chromium solution (Na251CrO4) (2 ml vial containing 2 mCi at a concentration of 1 mCi/ml) (PerkinElmer, catalog number: NEZ030S002MC )
- 1 M HCl (Sigma-Aldrich, catalog number: 258148 or equivalent )
Equipment
- Multichannel pipette, 8 or 12 channels [e.g., Finnpipette F2 10-100 μl, 8 channels (Sigma-Aldrich, catalog number: Z678007 )]
- Water bath
- Incubator 37 °C, 5% CO2
- Standard laboratory desktop centrifuge for 15 ml sample tubes and 96-well plates
- Packard TopCount NXT reader (Perkin Elmer, USA)
Software
- Prism
Procedure
- Cytotoxic T Lymphocyte (CTL) Assay with peptide titration at an E:T ratio of 10:1
This first section describes the different steps needed to perform a cytotoxic T lymphocyte (CTL) assay. Radioactive labelling of target cells with 51Cr (step A1). Preparation of the peptide dilution (step A2). Preparation of the effector CD8 T cells (step A3). Loading procedure into 96 wells plate (step A4). Determination of 51Cr radioactivity (step A5). Analysis of results (step A6).- Radioactive labelling of T2 target cells with Chromium 51 (51Cr)
- Calculate the number of T2 target cells needed for the CTL assay. You will need 1,000 target cells for 1 well of a 96-well plate. Consider adding 20% more cells to account for volume loss during pipetting.
Note: We usually label 0.5 x 106 T2 cells, which is enough for 500 wells. - Harvest the correct number of T2 target cells in 15 ml Falcon tubes and spin them for 5 min at 530 x g at room temperature (RT).
- Wash the cells in 1 ml RT RPMI/10% FBS medium.
- Spin down, remove all liquid and incubate T2 cells with 50 μl of pure 51Cr solution (activity 1 mCi/ml) for 45 min at 37 °C in a water bath. Take the necessary precaution and protection when manipulating radioactive material (e.g., lead shields, rapid manipulation). Contact your radiosafety officer for detailed procedures.
- At the end of 51Cr labeling time, add 10 ml of RPMI/10%FBS and spin 5 min at 530 x g. Discard the supernatant with the required safety precautions (contact your radiosafety officer for details).
- Repeat the washing step with 10 ml of RPMI/10%FBS two more times.
- After the third wash, resuspend the cells at a concentration of 0.02 x 106/ml in RT RPMI/10% FBS.
- 50 μl (1,000 cells) of freshly labelled 51Cr cell solution is added at the last step (see step A4) in each well of a 96-V-bottom plate.
- Preparation of the peptide dilutions
- Dissolve the antigenic peptide of interest [here NY-ESO-1157-165 peptide (SLLMWITQC)] in PBS to prepare a stock solution of 1mg/ml (~1 mM) for a 10-AA peptide.
- From the stock solution, prepare a 500x dilution in RPMI/10% FBS to get a starting concentration of 2 μM for the following serial dilution steps. This represents a 2x concentrated solution of antigenic peptide, given the 1:2 dilution step occurring during the plating step (step A4).
Note: To calculate the volume of antigenic peptide solution required for the serial dilutions, consider that each condition is tested in duplicate and that spare volume is needed. 200 μl [100 μl peptide per well x 2 (duplicates)] of peptide solution are required per T cell tested. Be sure to have at least 20% spare volume. Example for 4 CD8 T cells: 4 x [100 μl peptide x 2(duplicates)] = 800 μl required + spare volume → prepare 1 ml of initial 2 μM solution. - Prepare 7 tubes (15 ml Falcon) with RPMI/10% FBS and make serial 10x-dilutions from the 2 μM peptide solution, making sure to thoroughly vortex the tube and to change the pipet tips each time.
Note: Example for 4 CD8 T cells: 100 μl (2 μM) + 900 μl RPMI/10% FBS → 100 μl (0.2 μM)+ 900 μl RPMI/10% FBS → 100 μl (0.02 μM) + 900 μl RPMI/10% FBS and so on. - Pipette 100 μl of the 2x concentrated peptide solution per well of a 96-V-bottom plate, starting from the bottom (lowest concentration) to the top (highest concentration) using a 12-multichannel pipette and a bassin. This will avoid changing the tips each time.
- For controls, prepare one solution without peptide and one with an irrelevant peptide (e.g., a class I Flu-specific peptide) at the highest concentration (2 μM) for each T cell clone to be tested.
- Dissolve the antigenic peptide of interest [here NY-ESO-1157-165 peptide (SLLMWITQC)] in PBS to prepare a stock solution of 1mg/ml (~1 mM) for a 10-AA peptide.
- Preparation of CD8 T cell clones (effector cells)
- Calculate the number of cells needed for the test:
For each T cell clone, you will need 10,000 x 2(duplicates) = 20,000 x 8 dilutions = 160,000 cells and 40,000 cells for the controls w/o peptide and irrelevant peptide, making a total of 0.2 x 106 cells for each T cell clone to be tested. - Accounting for volume loss, prepare 0.25 x 106 T cells and resuspend them in 1.25 ml RT RPMI/10% FBS (0.2 x 106 cells/ml).
- Add 50 μl of the prepared effector cells (= 10,000 cells/well) to each well that already contains 100 μl of the serial peptide dilutions.
- Calculate the number of cells needed for the test:
- Steps to load the samples in assay plates (Figure 1)
- 1st Step: Peptide dilutions (100 μl) (step A2)
- 2nd Step: CD8 T cell clones (10,000 cells in 50 μl) (step A3)
- 3rd Step: 51Cr-labelled T2 (1,000 cells in 50 μl) (step A1)
- Controls: Spontaneous Release (SR): 4 wells with 150 μl of medium and 50 μl 51Cr- labelled T2. Total Release (TR): 4 wells with 150 μl of 1 M HCl and 50 μl 51Cr-labelled T2.
- When the plate is filled, incubate it for 4 h at 37 °C in an incubator with 5% CO2.
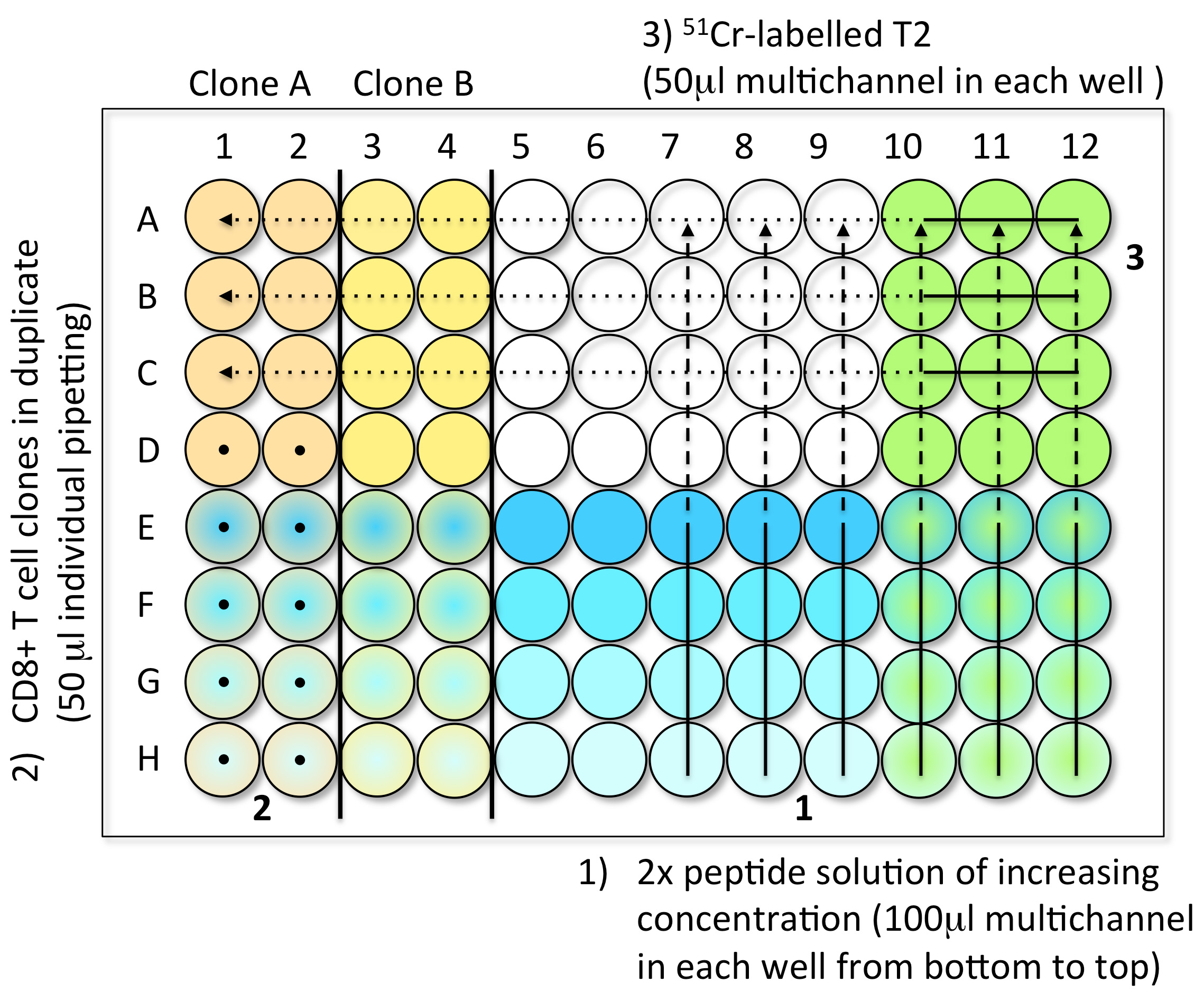
Figure 1. Steps to load the CTL test samples into V-bottom 96-well plates. Step 1: Load 100 μl of the 2x peptide solution with a multichannel pipette, from low concentration (bottom, light blue) to high concentration (top, dark blue). Step 2: Individually add 50 μl of effector CD8 T cell clones in duplicate (here clones A and B), from bottom to top (black dots). You can test up to six clones per 96-well plate. Step 3: Load 50 μl of freshly 51Cr-labelled target cells into every well using a multichannel pipette. The control wells are prepared in the last plate of the experiment. Avoid touching the liquid present in the wells while introducing the different components of the test. - 1st Step: Peptide dilutions (100 μl) (step A2)
- Determination of 51Cr radioactivity
At the end of the 4 h incubation, spin the plate for 2 min at 234 x g and transfer 50 μl of the supernatant from each well to a LumaPlate-96 using a multi-channel pipette. The amount of radiation emitted by 51Cr present in each supernatant is determined as Counts per Minute (CPM) using a TopCount NXT reader. - Calculation of specific lysis: EC50 and EMax
To calculate the amount of specific lysis that occurred in each well, use the following formula: 100x [experimental counts per minutes (cpm) - SR (average cpm)/(TR(average cpm) - SR (average cpm)] = % specific lysis.
The antigen sensitivity and maximum killing capacity of each clone can be quantified as EC50 and EMax, respectively. EC50 is defined by the peptide concentration producing half maximal lysis of target cells and can be derived by fitting a dose response curve analysis using the Prism software for example (Figure 2). EMax is defined as the relative maximal lysis capacity of the effector T cells (Figure 2).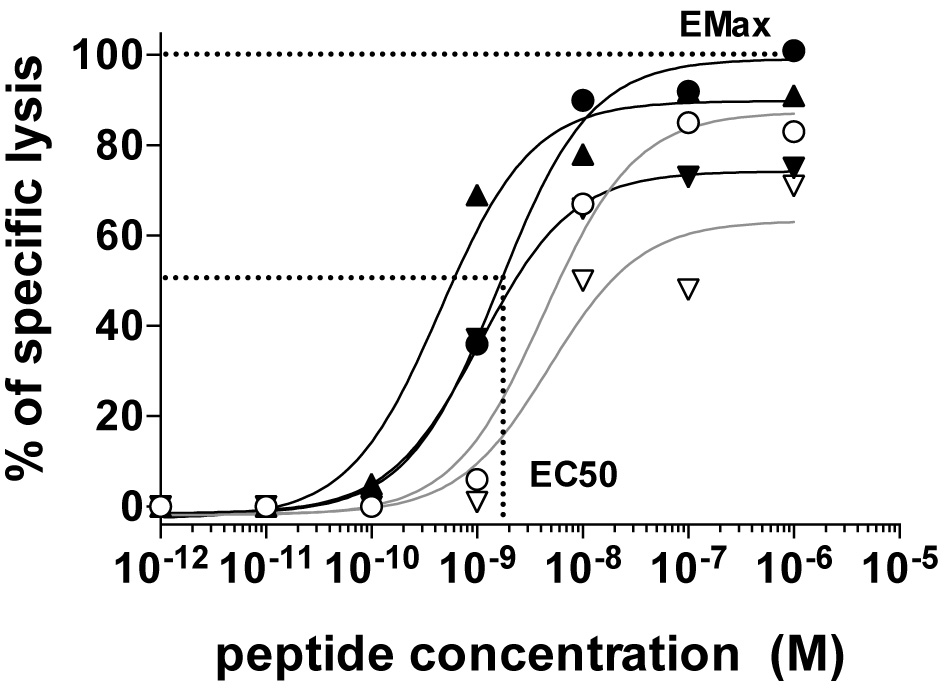
Figure 2. The relative functional avidity of five NY-ESO-1157-165-reactive CD8 T cell clones expressing distinct αβTCRs assessed by measuring the lysis capacity of T2 target cells pulsed with graded concentrations of the natural NY-ESO-1157-165 peptide (SLLMWITQC). Also shown is an example of EC50 (i.e., the peptide concentration required to achieve 50% of maximal lysis) and EMax [i.e., maximal lysis at high (saturating) antigen dose)] for the clone producing highest maximal lysis (black round). Details can be found in Hebeisen et al., 2015.
- Tumor killing (TK) Assay with tumor target cell titration
The following section describes the different steps needed to perform a tumor killing (TK) assay: Radioactive labelling of tumor target cells with 51Cr (step B1). Preparation of the effector CD8 T cells (step B2). Preparation of the peptide solutions (step B3). Loading procedure into 96-well plate (step B4). Determination of 51Cr radioactivity (step B5). Analysis of results (step B6).- Radioactive labeling of tumor cell line with 51Cr
- Calculate the number of target cells (e.g., Melanoma cell line Me275) needed for the TK assay. You will need 1,000 target cells for 1 well of a 96-well plate. Consider adding 20% more cells to account for volume loss during pipetting.
Note: We usually label 0.5 x 106 target cells, which is enough for 500 wells. - Harvest the correct number of tumor target cells in 15 ml Falcon tubes and spin them for 5 min at 530 x g at room temperature (RT).
- Wash the cells in 1 ml RT RPMI/10% FBS medium.
- Spin down, remove all liquid and incubate the tumor cells with 50 μl of pure 51Cr solution (activity 1 mCi/ml) for 45 min at 37 °C in a water bath.
- After the 51Cr labeling, add 10 ml of RPMI/10% FBS and spin 5 min at 530 x g. Discard the supernatant with the required safety precautions (contact your radiosafety officer for detailed procedures).
- Repeat the washing step with 10 ml of RPMI/10%FBS two more times.
- After the third wash, resuspend the cells at a concentration of 0.02 x 106/ml in RT RPMI/10% FBS.
- 50 μl (1,000 cells) of this cell solution is added at the last step (see step B4) in each well of a 96-V-bottom plate.
- Calculate the number of target cells (e.g., Melanoma cell line Me275) needed for the TK assay. You will need 1,000 target cells for 1 well of a 96-well plate. Consider adding 20% more cells to account for volume loss during pipetting.
- Preparation of T cell clones
- For the tumor killing assay done in duplicate, 0.1 x 106 T cells are needed per clone. If the test includes conditions with and without peptide pulsing on the target (see step B4), a minimum of 0.2 x 106 T cells are needed.
- CD8 T cell clones are counted and resuspended at a concentration of 0.6 x 106/ml in RT RPMI/10% FBS.
- From that solution, prepare a serial dilution directly in 96-V-bottom plates as follows:
Add 75 μl (45,000 cells) in the first well for the first condition [effector (E) to target (T) ratio 30:1] and 50 μl of RPMI/10% FBS in the three wells located underneath. Remember to do it twice for the duplicate. Proceed by transferring 25 μl of the first well in the second, mix well by pipetting up and down, and proceed likewise for the successive dilutions. At the end, all the wells contain 50 ul of medium with the number of cells indicated in brackets.
Note: A multi-channel pipet can be used to proceed entire 96-well plates rapidly.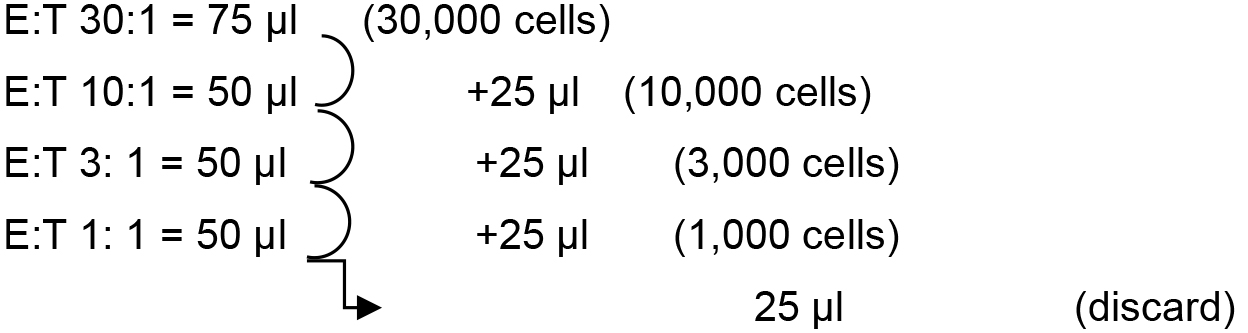
- For the tumor killing assay done in duplicate, 0.1 x 106 T cells are needed per clone. If the test includes conditions with and without peptide pulsing on the target (see step B4), a minimum of 0.2 x 106 T cells are needed.
- Peptide solution
- Dilute peptide stock solution (1 mM) in RPMI/10% FBS medium to make a 2x concentrated 2 μM solution. Final concentration per well with peptide will be 1 μM. For example, put 10 μl peptide stock solution (1 mM) in 5 ml RPMI/10% FBS (enough for 500 tests).
- Add either 100 μl RPMI/10% FBS (condition without peptide) or 100 μl 2x concentrated peptide solution to the appropriate wells.
Note: Per 96-well plate, we prepare the “no peptide” condition in the top 4 lines (A to D) and a “+ peptide” condition in the 4 bottom lines (E to H).
- Dilute peptide stock solution (1 mM) in RPMI/10% FBS medium to make a 2x concentrated 2 μM solution. Final concentration per well with peptide will be 1 μM. For example, put 10 μl peptide stock solution (1 mM) in 5 ml RPMI/10% FBS (enough for 500 tests).
- Steps to load the samples in assay plates (Figure 3)
- 1st Step: CD8 T cell clones serial dilution (50 μl) (step B2)
- 2nd Step: Solution with or without peptide (100 μl) (step B3)
- 3rd Step: 51Cr-labelled Me275 (1,000 cells in 50 μl) (step B1)
- Controls: Spontaneous Release (SR): 4 well with 150 μl of medium and 50 μl 51Cr-labelled T2 .Total Release (TR): 4 well with 150 μl of 1 M HCl and 50 μl 51Cr-labelled T2
- When the plate is filled, incubate the test samples for 4 h at 37 °C in an incubator with 5% CO2.
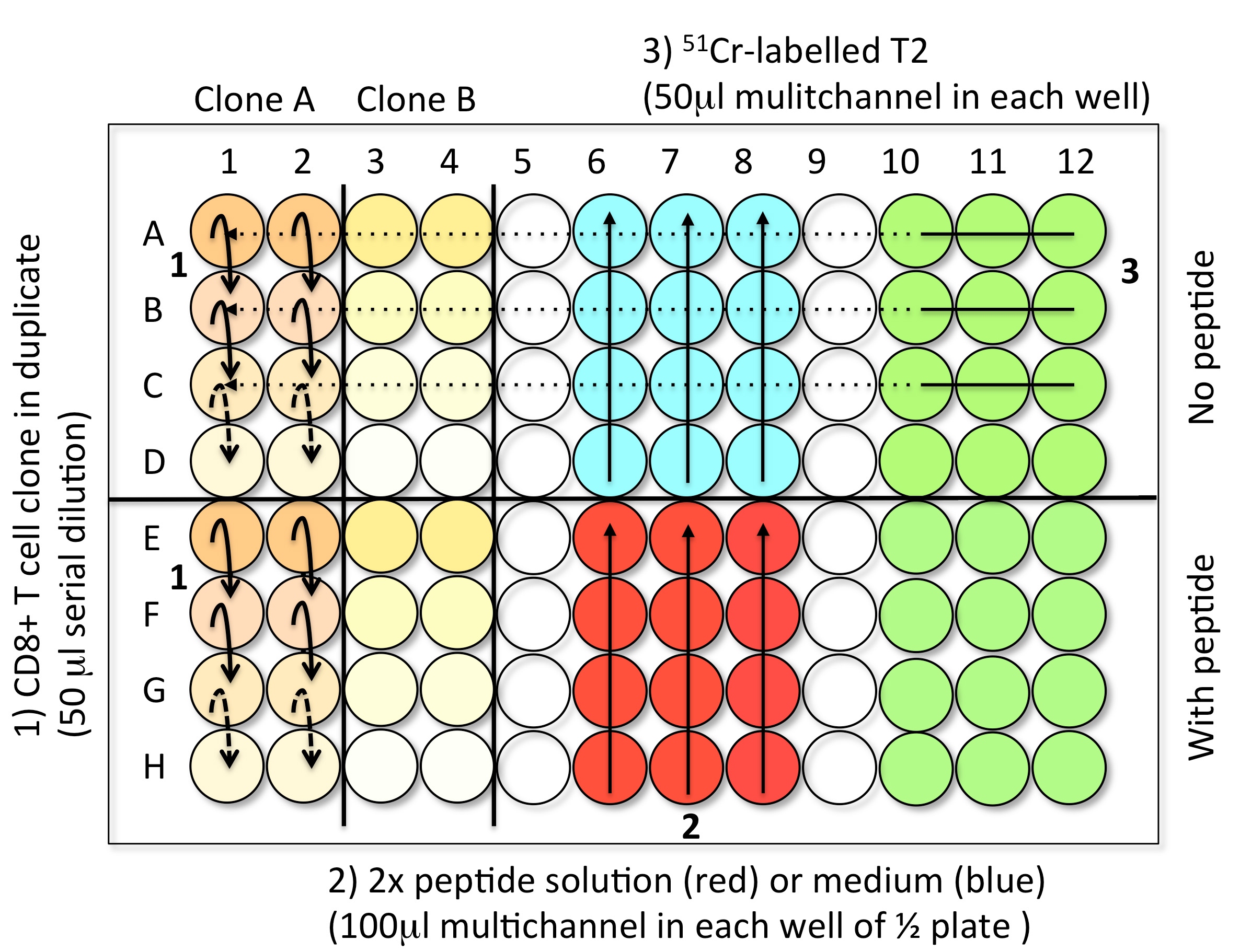
Figure 3. Steps to load the TK test samples into V-bottom 96-well plates. Step 1: Perform the serial dilution of CD8 T cell clones to get 50 μl of cells in each wells. For each clone (here A and B), reproduce the serial dilutions in the two conditions, with or without peptide. Step 2: Add 100 μl of 2x peptide solution (red) or medium (blue) in the corresponding halves of the plate using a multichannel pipette. Step 3: Load 50 μl of freshly 51Cr-labelled target cells into every well using a multichannel pipette. The control wells are prepared in the last plate of the experiment. Avoid touching the liquid present in the wells while introducing the different components of the test. - 1st Step: CD8 T cell clones serial dilution (50 μl) (step B2)
- Determination of 51Cr radioactivity
At the end of the 4 h incubation, spin the plate for 2 min at 234 x g and transfer 50 μl of the supernatant from each well to a LumaPlate-96 using a multi-channel pipette. The amount of radiation emitted by 51Cr present in each supernatant is determined as Counts per Minute (CPM) using a TopCount NXT reader. - Calculation of specific lysis
To calculate the amount of specific lysis that occurred in each well, use the following formula: 100x [(experimental counts per minutes (cpm) - SR (average cpm)/(TR (average cpm) - SR (average cpm)] = % specific lysis.
The efficacy by which the effector CD8 T cell recognize and lyse tumor cells at the distinct effector to target ratios can be represented as shown in Figure 4 by using the Prism software.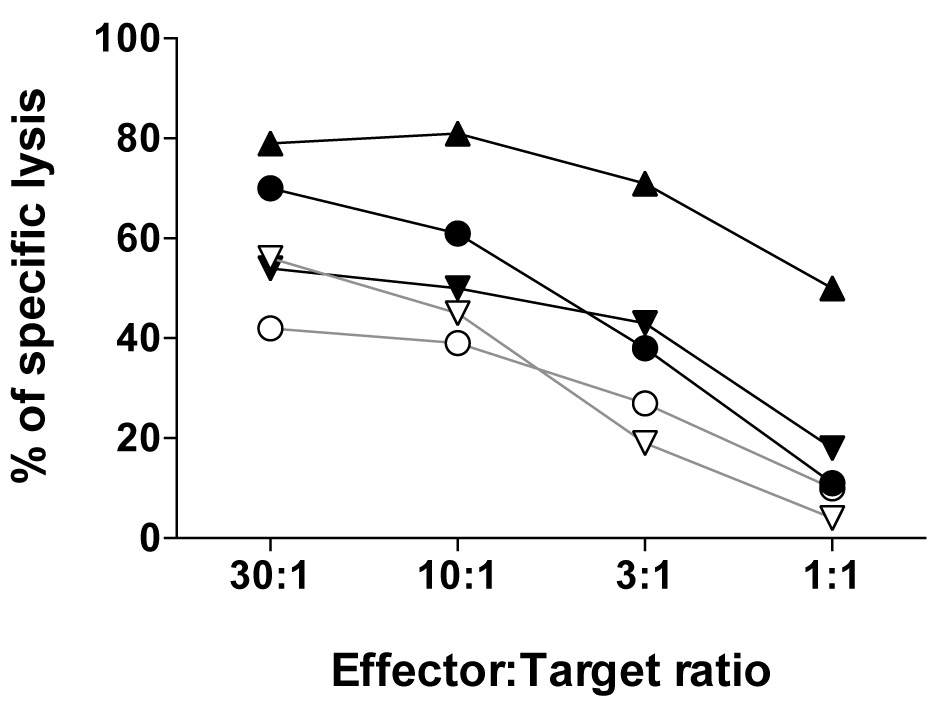
Figure 4. The relative tumor cell lysis capacity of five NY-ESO-1157-165-reactive CD8 T cell clones expressing distinct αβTCRs assessed using the melanoma cell line Me275 (A2pos/NY-ESO-1157-165pos) at the indicated effector:target ratios. For details, see Hebeisen et al., 2015.
- Radioactive labeling of tumor cell line with 51Cr
Notes
- It is very important to have effector and target cells in very good conditions (Rubio-Godoy et al., 2001). Effector cells should be in a resting phase (around D12-D16 post-restimulation) to avoid high unspecific background killing. Target cells in bad condition (dying or dead) cannot be efficiently labelled with chromium, which leads to results that are difficult to interpret.
- Each new unknown target cell should be tested first for its capability to be labelled with chromium (4 h spontaneous and total release only).
Acknowledgments
This work was supported by the Department of Oncology of the University of Lausanne and the Ludwig Center for Cancer Research of the University of Lausanne.
The Chromium Release Assay has been developed by researchers of the Swiss Institute of Experimental Cancer Research (ISREC) and the Department of Biochemistry at the University of Lausanne (Switzerland) and was first published in 1968 (see Reference 1).
References
- Brunner, K. T., Mauel, J., Cerottini, J. C. and Chapuis, B. (1968). Quantitative assay of the lytic action of immune lymphoid cells on 51-Cr-labelled allogeneic target cells in vitro; inhibition by isoantibody and by drugs. Immunology 14(2): 181-196.
- Hebeisen, M., Schmidt, J., Guillaume, P., Baumgaertner, P., Speiser, D. E., Luescher, I. and Rufer, N. (2015). Identification of rare high-avidity, tumor-reactive CD8+ T cells by monomeric TCR-ligand off-rates measurements on living cells. Cancer Res 75(10): 1983-1991.
- Rubio-Godoy, V., Dutoit, V., Rimoldi, D., Lienard, D., Lejeune, F., Speiser, D., Guillaume, P., Cerottini, J. C., Romero, P. and Valmori, D. (2001). Discrepancy between ELISPOT IFN-gamma secretion and binding of A2/peptide multimers to TCR reveals interclonal dissociation of CTL effector function from TCR-peptide/MHC complexes half-life. Proc Natl Acad Sci U S A 98(18): 10302-10307.
- Speiser, D. E., Baumgaertner, P., Voelter, V., Devevre, E., Barbey, C., Rufer, N. and Romero, P. (2008). Unmodified self antigen triggers human CD8 T cells with stronger tumor reactivity than altered antigen. Proc Natl Acad Sci U S A 105(10): 3849-3854.
- Zippelius, A., Pittet M. J., Batard P., Rufer N., de Smedt M., Guillaume P., Ellefsen K., Valmori D., Liénard D., Plum J., MacDonald H. R., Speiser D. E., Cerottini J.C. and Romero P. (2002). Thymic selection generates a large T cell pool recognizing a self-peptide in humans. J Exp Med 195(4): 485-94.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Baumgaertner, P., Speiser, D. E., Romero, P., Rufer, N. and Hebeisen, M. (2016). Chromium-51 (51Cr) Release Assay to Assess Human T Cells for Functional Avidity and Tumor Cell Recognition. Bio-protocol 6(16): e1906. DOI: 10.21769/BioProtoc.1906.
Category
Immunology > Immune cell function > Cytotoxicity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link













