- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Mouse Liver Mitochondria Isolation, Size Fractionation, and Real-time MOMP Measurement
Published: Vol 6, Iss 15, Aug 5, 2016 DOI: 10.21769/BioProtoc.1892 Views: 13025
Reviewed by: Pia GiovannelliAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of Pure Mitochondria from Rat Kidneys and Western Blot of Mitochondrial Respiratory Chain Complexes
Tamara Micakovic [...] Sigrid Christa Hoffmann
Oct 5, 2019 11364 Views

Protein Import Assay into Mitochondria Isolated from Human Cells
Lena M. Murschall [...] Jan Riemer
Jun 20, 2021 6329 Views

Analysis and Quantification of the Mitochondrial–ER Lipidome
Alexis Diaz-Vegas [...] James G. Burchfield
Jul 5, 2024 2587 Views
Abstract
The mitochondrial pathway of apoptosis involves a complex interplay between dozens of proteins and lipids, and is also dependent on the shape and size of mitochondria. The use of cellular models in past studies has not been ideal for investigating how the complex multi-factor interplay regulates the molecular mechanisms of mitochondrial outer membrane permeabilization (MOMP). Isolated systems have proven to be a paradigm to deconstruct MOMP into individual steps and to study the behavior of each subset of MOMP regulators. In particular, isolated mitochondria are key to in vitro studies of the BCL-2 family proteins, a complex family of pro-survival and pro-apoptotic proteins that directly control the mitochondrial pathway of apoptosis (Renault et al., 2013).
In this protocol, we describe three complementary procedures for investigating in real-time the effects of MOMP regulators using isolated mitochondria. The first procedure is “Liver mitochondria isolation” in which the liver is dissected from mice to obtain mitochondria. “Mitochondria labeling with JC-1 and size fractionation” is the second procedure that describes a method to label, fractionate by size and standardize subpopulations of mitochondria. Finally, the “Real-time MOMP measurements” protocol allows to follow MOMP in real-time on isolated mitochondria. The aforementioned procedures were used to determine in vitro the role of mitochondrial membrane shape at the level of isolated cells and isolated mitochondria (Renault et al., 2015).
Materials and Reagents
- 50 ml conical centrifuge tube (Santa Cruz Biotechnology, catalog number: sc-200251 )
- Petri dish, 100 x 15 mm (Fisher Scientific, catalog number: FB0875712 )
- 15 ml conical centrifuge tube (Santa Cruz Biotechnology, catalog number: sc-200250 )
- 1.5 ml Micro centrifuge tube (USA Scientific, catalog number: 1615-5510 )
- Gravity chromatography column (Thermo Fisher Scientific, PierceTM, catalog number: 29920 )
- Pasteur pipet (Santa Cruz Biotechnology, catalog number: sc-204537 )
- 96-well plate, flat bottom, black polystyrene (Corning, CostarTM, catalog number: 3915 )
- C57BL/6 mice (Charles River Laboratories, catalog number: 027 )
- 1x phosphate buffered saline (PBS), pH 7.4 (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) (Fisher Scientific, catalog number: BP24384 )
- Trehalose (Sigma-Aldrich, catalog number: 1673715 )
- Sucrose (Sigma-Aldrich, catalog number: S0389 )
- [4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid]-KOH (HEPES) (Sigma-Aldrich, catalog number: H0527 )
- KCl (Sigma-Aldrich, catalog number: P9541 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: E9884 )
- Ethyleneglycoltetraacetic acid (EGTA) (Sigma-Aldrich, catalog number: E3889 )
- Bovine serum albumin-fraction V (Sigma-Aldrich, catalog number: A9418 )
- Protease inhibitor cocktail (Thermo Fisher Scientific, HALTTM, catalog number: 78430 )
- Sepharose CL-2B resin (Sigma-Aldrich, catalog number: CL2B300 )
- 5,5’,6,6’-tetrachloro-1,1’,3,3’-tetraethylbenzimidazolylcarbocyanine iodide (JC-1) (Thermo Fisher Scientific, catalog number: T3168 )
- Triton X-100 (Fisher Scientific, catalog number: BP151-500 )
- Carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP) (Sigma-Aldrich, catalog number: C2920 )
- Recombinant BAX [purified using the IMPACT (Intein Mediated Purification with an Affinity Chitin-binding Tag) system (New England Biolabs, catalog number: E6901S )]
- β-octylglucoside (OG) (Sigma-Aldrich, catalog number: O8001 )
- Dimethylsulfoxide (DMSO) (Fisher Scientific, catalog number: BP231-100 )
- Magnesium Chloride Hexahydrate (Fisher Scientific, catalog number: M33-500 )
- Trehalose isolation buffer (TIB) (see Recipes)
- JC-1 loading buffer (see Recipes)
- Liposome buffer (see Recipes)
Equipment
- Dissection tools (scissors, scalpel, forceps)
- Razor blade (Daigger Scientific, catalog number: EF7281A )
- 15 ml Potter-Elvehjem dounce homogenizer (Omni International, catalog number: 07-358044 )
- Swing bucket centrifuge (Thermo Fisher Scientific, Thermo ScientificTM SorvallTM, model: Legend XTR )
- Spectrophotometer (Fisher Scientific, GE Healthcare UltrospecTM, model: Ultrospec 7000 )
- Water bath (Fisher Scientific, IsoTempTM, model: 215 )
- Tabletop centrifuge (Thermo Fisher Scientific, SorvallTM LegendTM, model: Micro 21 )
- Fluorescence plate reader (Biotek, model: Synergy H1 )
Procedure
- Liver mitochondria isolation
Animal handling, euthanasia, and dissection must be done following the Institutional Animal Care and Use Committee guidelines. We have successfully utilized carbon dioxide asphyxiation before cervical dislocation to euthanize the mice. However, we advise the reader to consult and follow the Animal Care and Use guidelines of their own institute for the appropriate procedure. Following steps involved in animal handling and dissection are executed wearing gloves.- Visualize the liver, the bile duct, and gallbladder. The bile duct and gallbladder must be removed or kept separate from the liver to avoid contamination of the mitochondria with bile (Figure 1a-c).
- Excise the four liver lobes and transfer immediately to a 50 ml conical tube containing ice-cold PBS. In case of blood contamination, soak the liver in ice-cold PBS until no more blood is washed out (Figure 1d-e).
Note: From this point, all the steps must be performed using reagents and materials at 4 °C to minimize the activation of proteases and phospholipases.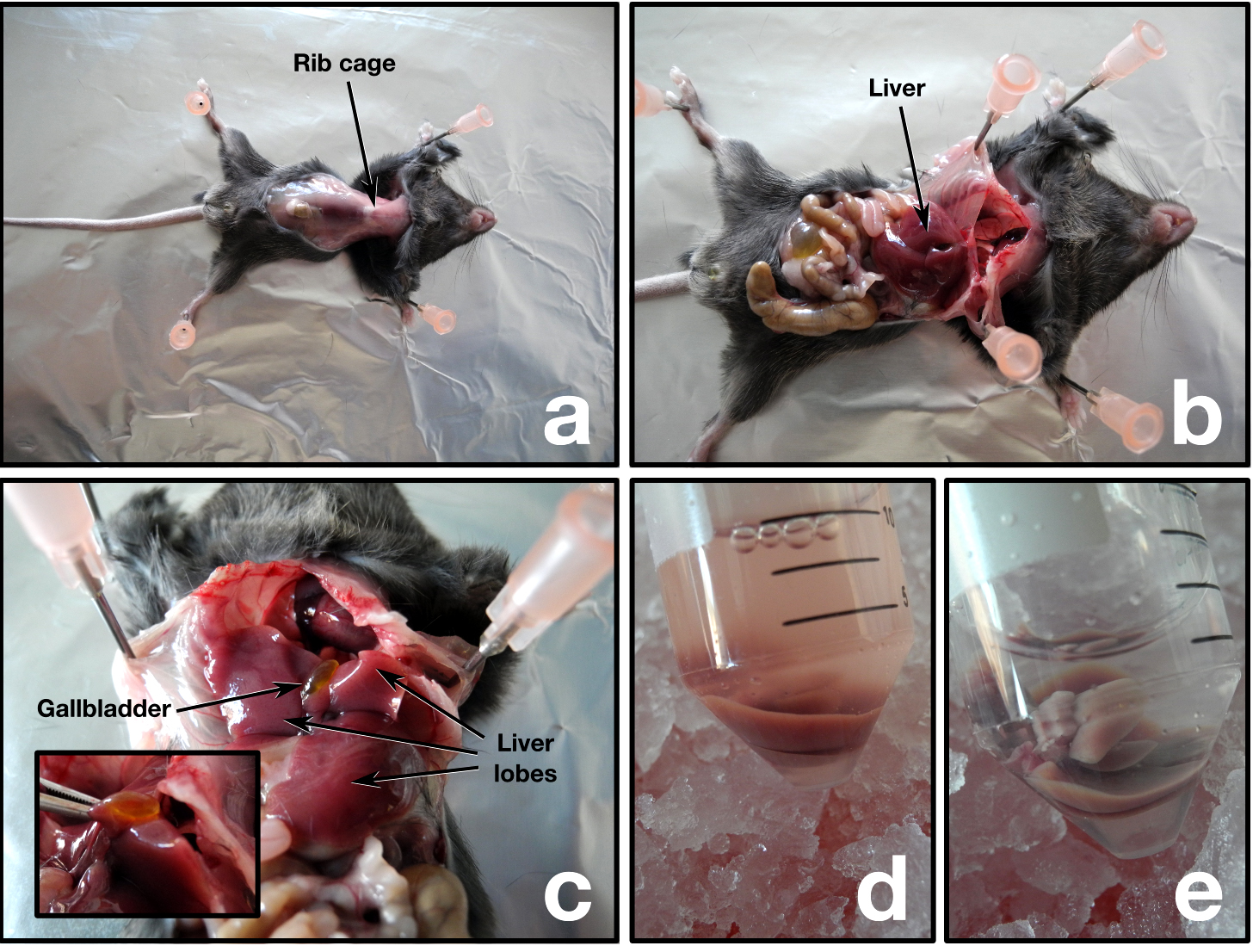
Figure 1. Mouse dissection - Transfer the liver with forceps to a clean Petri dish placed on ice and drain off the excess PBS.
- Mince the liver using a clean razor blade until forming a homogeneous paste (Figure 2).
- Transfer half of the liver paste using the razor blade as a spoon into a chilled Potter-Elvehjem dounce homogenizer containing 10 ml TIB.
- Insert the pestle into the homogenizer and gently push it downwards. Early resistance is common during the first strokes, and no excessive force should be applied to the liver paste. Instead, move the pestle upwards to resuspend the liver paste and slowly repeat those steps until all the material is able to go past the pestle without strong resistance. The homogenization step can be done on the bench as long the whole procedure is done quickly enough to maintain the temperature close to 4 °C.
- Homogenize the paste five times, pour into a 15 ml conical tube and store on ice until all the liver paste is processed (Figure 2).
- Repeat steps 6-8 with the second half of the liver paste.
- The following differential centrifugation steps are required to obtain the mitochondrial fraction. All the steps should be performed in a swinging bucket centrifuge at 4 °C. Resuspension of pellets is done gently by pipetting using a P1000 pipette (Figure 2).
- 600 x g for 10 min. Transfer the supernatant (S/N) to a clean, pre-chilled 15 ml conical tube.
- 3,500 x g for 10 min. Discard the S/N. Resuspend the pellet in 10 ml TIB.
- 1,500 x g for 5 min. Transfer the S/N to a clean, pre-chilled 15 ml conical tube.
- 5,500 x g for 10 min. Discard the S/N. Resuspend the pellet in 10 ml TIB.
- Repeat steps A9b to A9d, but resuspend the final pellet in 500 μl TIB.
- 600 x g for 10 min. Transfer the supernatant (S/N) to a clean, pre-chilled 15 ml conical tube.
- To quantify the amount of isolated material in the resuspended pellet (step A9e), add 5 μl of the resuspended pellet to 995 μl of TIB (1:200 dilution) and measure the OD520 with a spectrophotometer. Dilute the mitochondria with TIB to standardize the sample to an OD520 value of 0.25 (~20 μg/μl of protein).
- At this point the isolated mitochondria can be aliquoted (50 µl), frozen in dry ice-ethanol to prevent formation of water crystals, and stored at -80 °C.
Note: Freshly isolated mitochondria from the previous step should be kept on ice and used within 2 h to ensure integrity of the outer membrane. If frozen mitochondria are used, thaw the sample in a 30 °C water bath and continue using TIB in the subsequent procedures.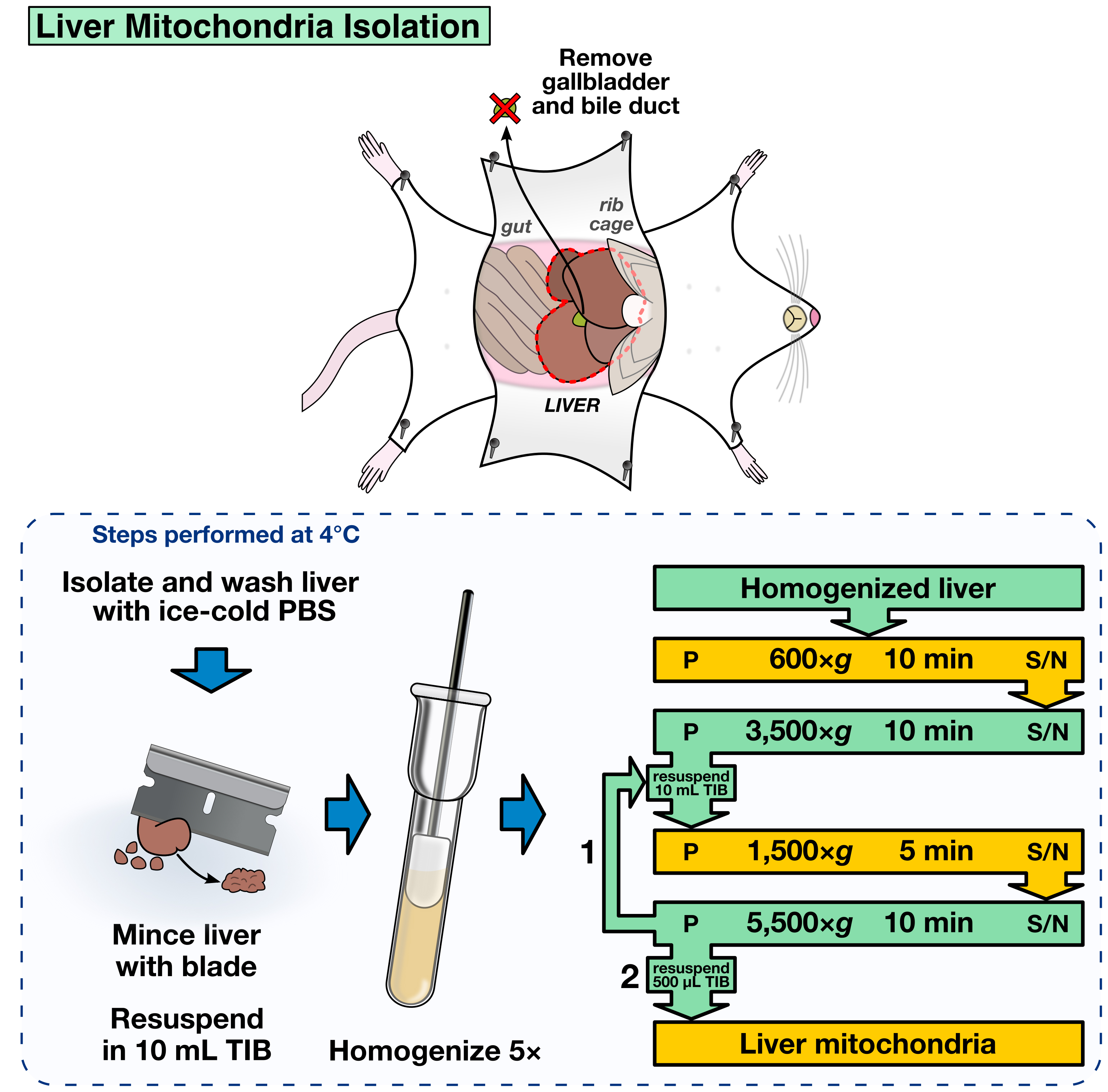
Figure 2. Liver mitochondria isolation
- Visualize the liver, the bile duct, and gallbladder. The bile duct and gallbladder must be removed or kept separate from the liver to avoid contamination of the mitochondria with bile (Figure 1a-c).
- Mitochondria labeling with JC-1 and size fractionation
Labeling mitochondria with JC-1, a potential-dependent mitochondrial dye that changes color as membrane potentials increase during MOMP, allows to standardize the mitochondria fractions and to record MOMP in real-time (as example, see Renault et al., 2015, Figure 4F). The labeling and fractionation step offers the opportunity to select the mitochondria by size: big mitochondria will be eluted prior to smaller ones. This fractionation step is optional and requires to calibrate the column using liposomes of a known size as a standard. Another option is to determine the average mitochondrial size in each fraction a posteriori, using for example dynamic light scattering. If fractionation by size is not desired, all the mitochondria-containing fractions can be combined in step B8.- Dilute 50 µl of mitochondria in 250 µl TIB buffer.
- Add JC-1 (200 µM) to the diluted mitochondria to a final concentration of 15 µM and incubate for 10 min at 30 °C (Figure 3).
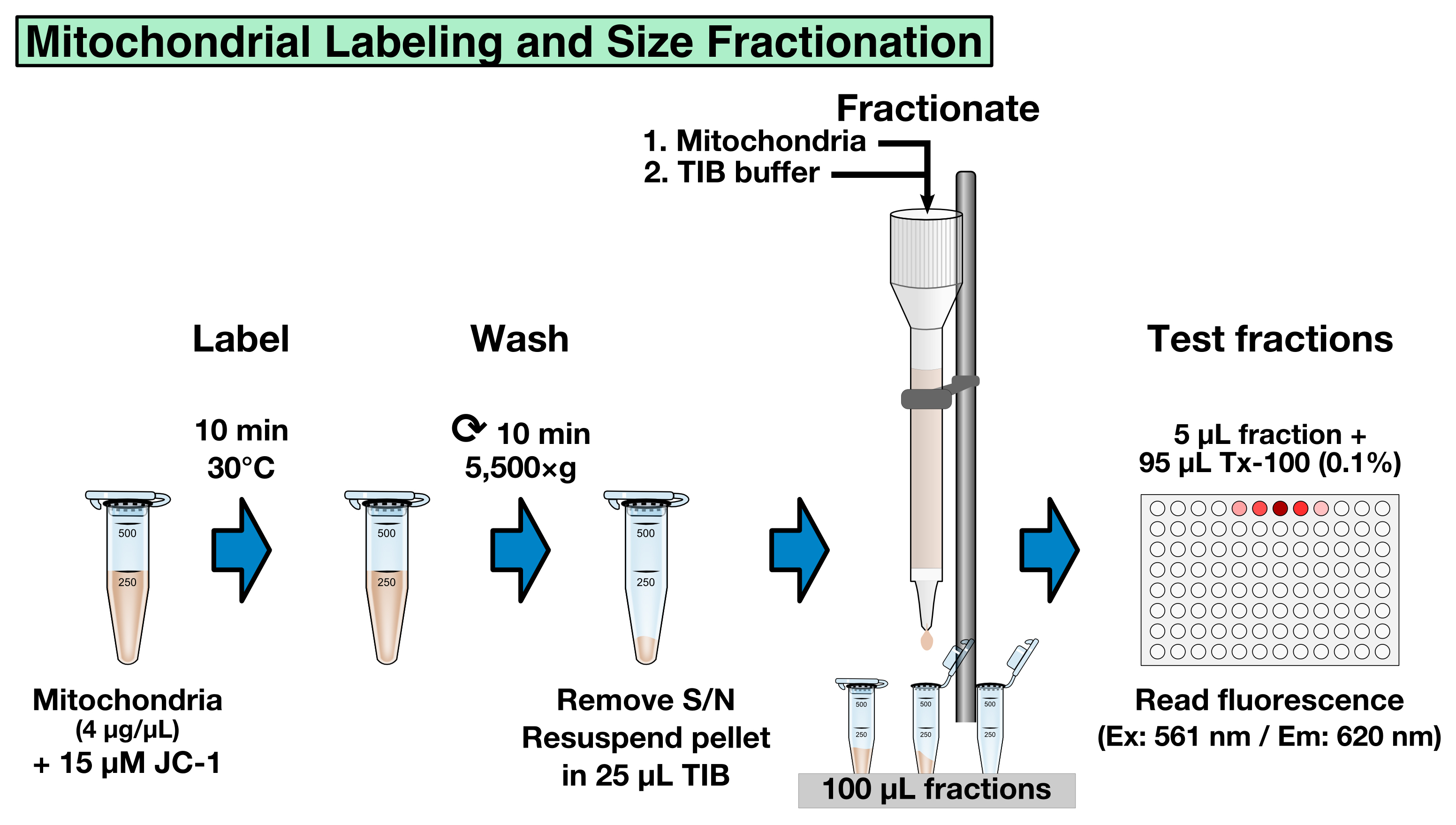
Figure 3. Mitochondrial labeling and size fractionation
- To remove unbound JC-1, centrifuge the mitochondria at 5,550 x g for 10 min at room temperature and resuspend the pellet in 25 µl TIB.
- Pre-equilibrate a 2 ml CL-2B gravity-column with 2 column volumes of TIB (4 ml) using gravity to let the TIB to flow through the CL-2B resin, load the resuspended JC-1 mitochondria onto the column, and allow sample to flow through.
Note: Make sure the resin does not run dry before loading the JC-1 labeled mitochondria. - Slowly apply 4 column volumes of TIB (4 x 2 ml) to the column using a clean Pasteur pipet and collect 20 x 100 µl (~2-3 drops) fractions in micro-centrifuge tubes.
- To determine which fractions contain labeled mitochondria, pre-fill a 96-well plate with 95 µl of 0.1% Triton X-100 in water (0.1% Triton X-100 permeabilizes mitochondria) and add 5 µl sample of each fraction to a separate well.
- Measure the fluorescence using a spectrophotometer (Ex: 561 nm/Em: 620 nm) to identify the mitochondria-containing fractions (typical values are 10,000-50,000 Relative Fluorescence Units for big mitochondria, and > 500 RFU for small size mitochondria).
- Combine the desired fractions containing JC-1 labeled mitochondria and standardize the samples using relative JC-1 fluorescence intensity (determine the fluorescence of each combined sample as described in step B6-7 and dilute the most concentrated samples with TIB buffer to reach the concentration of the lowest sample).
Note: In our experience 0.05-1 µm liposomes were subjected onto the CL-2B gravity-column and used as reference for selecting appropriate fractions. Briefly, fluorescent liposomes of several sizes (0.05, 0.2, and 1 µm) were prepared according to Asciolla et al., 2012. The liposomes were loaded onto the CL-2B gravity column and 100 µl fractions were collected, as described in steps B4-6 of this protocol. Identification of liposome-containing fractions for each liposome size allows to determine in which fractions vesicles of a given size should be expected.
- Dilute 50 µl of mitochondria in 250 µl TIB buffer.
- Real-time MOMP measurements
Real-time MOMP quantification is determined by measuring the mitochondrial membrane potential (ΔΨM) using the fluorescent JC-1 dye. The JC-1 dye exhibits potential-dependent accumulation in mitochondria, indicated by a fluorescence emission shift from green (~529 nm) to red (~590 nm). MOMP and the subsequent loss of ΔΨM are indicated by a decrease in the red/green fluorescence intensity ratio. There is a plethora of options to investigate a subset of inducers and regulators of MOMP in real-time. It is important to have internal and experimental controls. An example is given in Table 1. For the positive internal control, we use FCCP that uncouples the electron transport from oxidative phosphorylation in mitochondria and completely depolarizes the mitochondrial membrane.
To generate a positive experimental control, we use detergent-activated recombinant BAX (Hsu and Youle, 1997). The detergent, β-octylglucoside (OG), artificially triggers BAX activation, and therefore BAXOG (20-50 nM) can be used as a reliable positive control (for 100 µl of 2.3 µM BAXOG: 5 µg BAX + 0.7% OG in liposome buffer, incubate for 60 min at 4 °C, aliquot and store at -80 °C).
Real-time measurements- Prepare a 2x proteins/inducers solution in TIB and dispense 50 µl in each well in a 96-well plate. The plate should contain the following internal and experimental controls (Figure 4; Table 1)
- Negative internal control: TIB only
- Positive internal control: FCCP (10 µM final concentration)
- Negative experimental control: purified recombinant BAX (20-50 nM final concentration)
- Positive experimental control: β-octylglucoside-activated BAX (BAXOG, 20-50 nM final concentration)
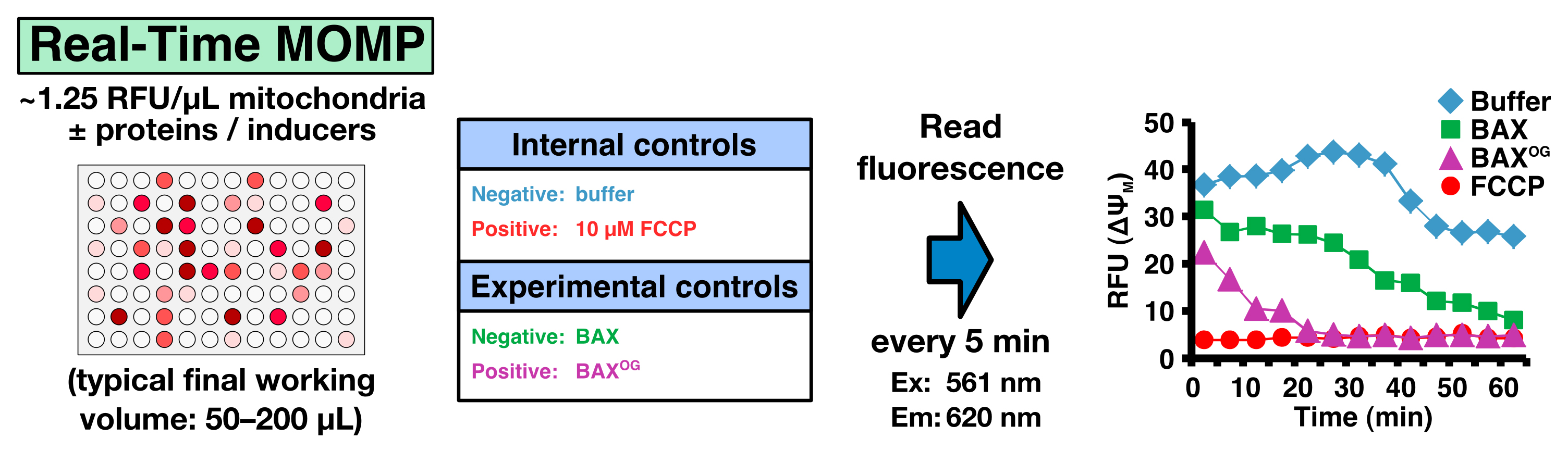
Figure 4. Real-Time MOMP - Negative internal control: TIB only
- Prepare a 2x suspension of JC-1 loaded mitochondria (~2.5 RFU/μl) and add 50 μl to each well to reach a final concentration of 1.25 RFU/μl.
Note: ~50-100 RFUs per reaction is ideal. - Measure fluorescence every 5 min in a plate reader for 60 min (Ex: 561 nm/Em: 620 nm) at 37 °C.
- Calculate the percentage of mitochondria that have undergone MOMP using the following formula:
% MOMP = (RFUbuffer − RFUsample) / (RFUbuffer − RFUFCCP)
Where RFUbuffer and RFUFCCP represent the fluorescence of the negative and positive internal controls, respectively, and RFUsample the fluorescence of the sample (see Figure 4G-H as example in Renault et al., 2015).
Table 1. An example to measure MOMP in real-time with internal and experimental controls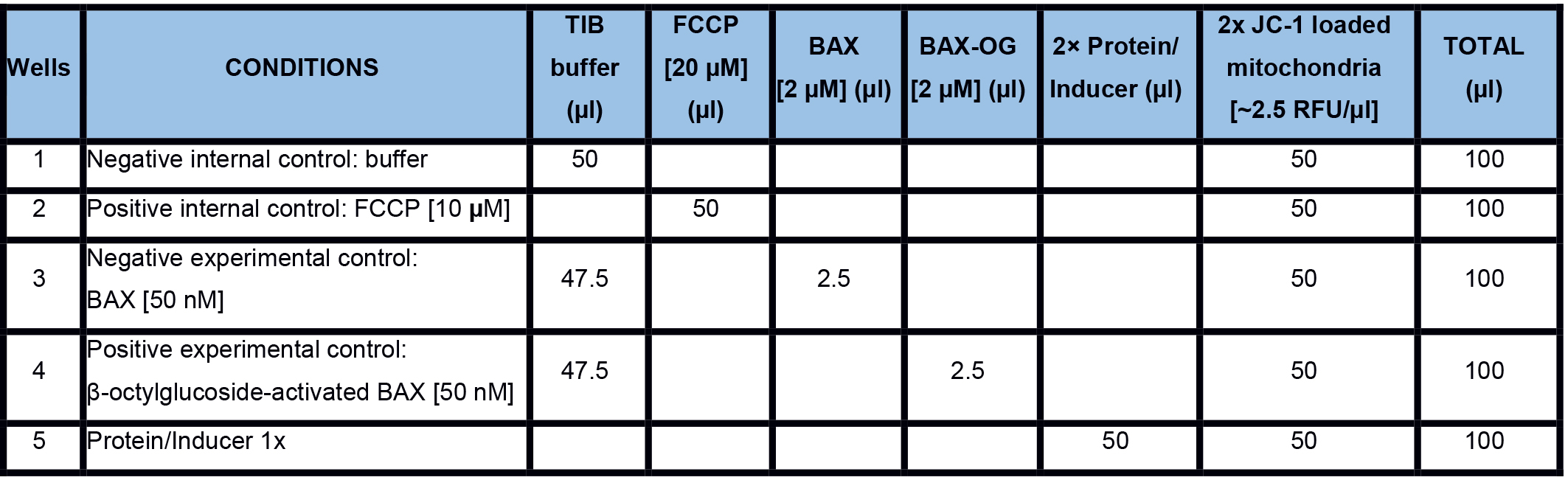
- Prepare a 2x proteins/inducers solution in TIB and dispense 50 µl in each well in a 96-well plate. The plate should contain the following internal and experimental controls (Figure 4; Table 1)
Recipes
- Trehalose isolation buffer (TIB)
200 mM trehalose
68 mM sucrose
10 mM HEPES-KOH, pH 7.4
10 mM KCl
1 mM EDTA
1 mM EGTA
0.1% BSA
Protease inhibitors cocktail (freshly added according to manufacturer's instructions)
Note: TIB should be prepared using BSA-Fraction V to eliminate fatty acid and lipid contaminants that promote non-specific BAK/BAX activation. - JC-1 loading buffer
200 μM stock solution is prepared in dimethylsulfoxide (DMSO) and diluted accordingly in TIB. - Liposome buffer
0.2 mM EDTA
10 mM HEPES-KOH, pH 7.4
200 mM KCl
5 mM MgCl2
Acknowledgments
We would like to thank everyone in the Chipuk Laboratory for their assistance and support. This work was supported by the following: NIH grants CA157740 and CA206005 (to J.E. Chipuk.); a pilot project from NIH P20AA017067 (to J.E. Chipuk), the JJR Foundation (to J.E. Chipuk), the William A. Spivak Fund (to J.E. Chipuk), the Fridolin Charitable Trust (to J.E. Chipuk), and an American Cancer Society Research Scholar Award (to J.E. Chipuk). This work was also supported in part by two research grants (5-FY11-74 and 1-FY13-416) from the March of Dimes Foundation (to J.E. Chipuk), the Leukemia and Lymphoma Scholar Award (to J.E. Chipuk), and the Developmental Research Pilot Project Program within the Department of Oncological Sciences at Mount Sinai (to J.E. Chipuk).
References
- Asciolla, J. J., Renault, T. T. and Chipuk, J. E. (2012). Examining BCL-2 family function with large unilamellar vesicles. J Vis Exp (68).
- Hsu, Y. T. and Youle, R. J. (1997). Nonionic detergents induce dimerization among members of the Bcl-2 family. J Biol Chem 272(21): 13829-13834.
- Renault, T. T., Floros, K. V. and Chipuk, J. E. (2013). BAK/BAX activation and cytochrome c release assays using isolated mitochondria. Methods 61(2): 146-155.
- Renault, T. T., Floros, K. V., Elkholi, R., Corrigan, K. A., Kushnareva, Y., Wieder, S. Y., Lindtner, C., Serasinghe, M. N., Asciolla, J. J., Buettner, C., Newmeyer, D. D. and Chipuk, J. E. (2015). Mitochondrial shape governs BAX-induced membrane permeabilization and apoptosis. Mol Cell 57(1): 69-82.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Renault, T. T., Luna-Vargas, M. P. and Chipuk, J. E. (2016). Mouse Liver Mitochondria Isolation, Size Fractionation, and Real-time MOMP Measurement. Bio-protocol 6(15): e1892. DOI: 10.21769/BioProtoc.1892.
Category
Cell Biology > Organelle isolation > Mitochondria
Cancer Biology > Cell death > Animal models
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











