- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
A Live-imaging, Heat Shock-inducible System to Measure Aux/IAA Degradation Rates in Planta
Published: Vol 6, Iss 15, Aug 5, 2016 DOI: 10.21769/BioProtoc.1881 Views: 10812
Reviewed by: Arsalan DaudiEunsook ParkStefanie Rosa

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Using a Live Analysis System to Study Amyloplast Replication in Arabidopsis Ovule Integuments
Makoto T. Fujiwara [...] Ryuuichi D. Itoh
Jun 5, 2025 2572 Views

Live Leaf-Section Imaging for Visualizing Intracellular Chloroplast Movement and Analyzing Cell–Cell Interactions
Yuta Kato [...] Mitsutaka Taniguchi
Aug 5, 2025 2322 Views

A Simple Protocol for Periodic Live Cell Observation of Flagellate Stages in the Lichen Alga Trebouxia
Enrico Boccato [...] Mauro Tretiach
Jan 20, 2026 188 Views
Abstract
An emerging theme in biology is the importance of cellular signaling dynamics. In addition to monitoring changes in absolute abundance of signaling molecules, many signal transduction pathways are sensitive to changes in temporal properties of signaling components (Purvis and Lahav, 2013). The phytohormone auxin regulates myriad processes in plant development. Many of these require the nuclear auxin signaling pathway, in which degradation of the Aux/IAA repressor proteins allows for transcription of auxin-responsive genes (Korasick et al., 2015). Using a heterologous yeast system, we found that Aux/IAAs exhibit a range of auxin-induced degradation rates when co-expressed in isolation with F-box proteins (Havens et al., 2012). Subsequent studies connecting signaling dynamics to plant growth and development confirmed that Aux/IAAs show similar differences in plants (Guseman et al., 2015; Moss et al., 2015). Here, we describe in detail the use of a heat-shock-inducible fluorescence degradation system to capture Aux/IAA degradation in real time in live plant roots. By employing this method, we were able to obtain high Aux/IAA expression and avoid the dampening long term effects of turnover, feedback and silencing. Degradation was dependent on the presence of an Aux/IAA degron and rates increased in response to exogenous auxin.
Keywords: AuxinMaterials and Reagents
- 1,000 μl pipet tip (Thermo Fisher Scientific, FisherbrandTM, catalog number: 02-681-4 )
- 50 ml conical centrifuge tube (Thermo Fisher Scientific, FisherbrandTM, catalog number: 06-443-20 )
- 1.5 ml microcentrifuge tubes (Thermo Fisher Scientific, FisherbrandTM, catalog number: 05-408-129 )
- Square (100 x 100 x 15 mm) Petri plates (Thermo Fisher Scientific, FisherbrandTM, catalog number: 08-757-11A )
- Round (35 mm) Petri plates (VWR International, catalog number: 10799-192 )
- Micropore tape 1530-0 (3M, catalog number: 70200412230 )
- Aluminum foil
- Razor blades
- Rectangular (24 x 50 mm) microscopy coverslips (Thermo Fisher Scientific, catalog number: 12-544E )
- Labeling tape
- > 4 ml glass perfume spray bottles (available in craft stores)
- Arabidopsis thaliana Columbia-0 (Col) seeds transformed with heat-shock constructs (Guseman et al., 2015; Moss et al., 2015)
- Bacto Agar (BD, catalog number: 214010 )
- Linsmaier-Skoog Media (Caisson Laboratories, catalog number: LSP03-1LT ) (see Note 1)
- Triton X-100 (GE Healthcare, catalog number: US22686 )
- Indole-3-acetic acid (IAA) (bioWORLD, catalog number: 705490 )
- Kanamycin (Thermo Fisher Scientific, catalog number: BP906-5 )
- 95% ethanol
- Seed sterilization solution (see Recipes)
- 0.5x Linsmaier-Skoog (LS) liquid media (see Recipes)
- Sterile plating agar solution (see Recipes)
- 0.5x LS media + 0.8% agar (see Recipes)
- Cover slip media (0.5x LS Media + 1.2% agar) (see Recipes)
- IAA stock solution (5 mM) (see Recipes)
Equipment
- Slide warmer (Thermo Fisher Scientific, model: 11-474-470 )
- Microscope (Leica Biosystems, model: DMI 3000B ) fitted with a Lumencor SOLA light source and YFP filter cube
- Leica long working 40x HCX PL FLUOTAR objective (see Note 2)
- Leica camera (Leica Microsystems, model: DFC345 FX )
- Forceps (Electron Microscopy Sciences, model: Dumont Tweezers Style 2A )
- Timer
Software
- Leica LAS AF version 2.6.0 for image acquisition
- Fiji/ImageJ software for image analysis
- GraphPad Prism6 software package for graphing and statistical analysis
Procedure
Note: Expression levels of Aux/IAA proteins are typically very low, so we have modified an existing heat-shock-inducible promoter system (Gray et al., 2001) to generate high levels of a fluorescently-tagged Aux/IAA for live cell imaging in root tissues prior to hormone treatment.
- Generation of transgenic Arabidopsis plants
- Heat-shock inducible VENUS-tagged Aux/IAA constructs (HS::VENUS-IAA-NLS) were cloned as previously described (Guseman et al., 2015; Moss et al., 2015) and based on pGREEN vectors containing the soybean heat shock promoter HS6871 (Gray et al., 2001).
- Arabidopsis plants were transformed with these constructs using the floral dip method (Clough and Bent, 1998). The plants were then selected for two generations on 0.5x LS agar plates containing 50 mg/ml kanamycin, to obtain T2 lines. For the experiments described below, 2-3 independent T2 lines for each construct should be used (see Notes 3-5).
- Heat-shock inducible VENUS-tagged Aux/IAA constructs (HS::VENUS-IAA-NLS) were cloned as previously described (Guseman et al., 2015; Moss et al., 2015) and based on pGREEN vectors containing the soybean heat shock promoter HS6871 (Gray et al., 2001).
- Plating and growing seedlings
Note: steps B1-4 below should be carried out in aseptic conditions in a sterile hood.- Pour 0.5x LS + 0.8% agar into square Petri plates in a sterile hood. Pour 1 plate for each independent plant line per experiment.
- Aliquot ~30 seeds into 1.5 ml tubes and add 300 μl of seed sterilization solution. Set tubes on their sides to ensure all seeds are in contact with solution. Let sit for 15 min and then remove sterilization solution and replace with 95% ethanol. Invert several times to wash. Completely remove 95% ethanol using a pipette and replace with sterile plating agar solution. Be sure to also plate non-fluorescent siblings (or other non-fluorescent line) to use as controls.
- Use a 1,000 μl pipette tip to gently aspirate seeds and transfer onto 0.5x LS plates. For each independent line, plate into 2 rows of 12 plants per plate (Figure 1A). Seal plates with micropore tape or Parafilm.
- Stratify seedlings vertically at 4 °C in the dark (cover with aluminum foil) for 2 days.
- Grow vertically in a growth chamber set under 16L:8D light conditions and 20 °C for 7 days.
- Pour 0.5x LS + 0.8% agar into square Petri plates in a sterile hood. Pour 1 plate for each independent plant line per experiment.
- Heat shock induction
- After 7 days of growth, place plates horizontally on a slide warmer set at 37 °C for 2 h to induce expression of the VENUS-IAA protein (Figures 1B and 2A).
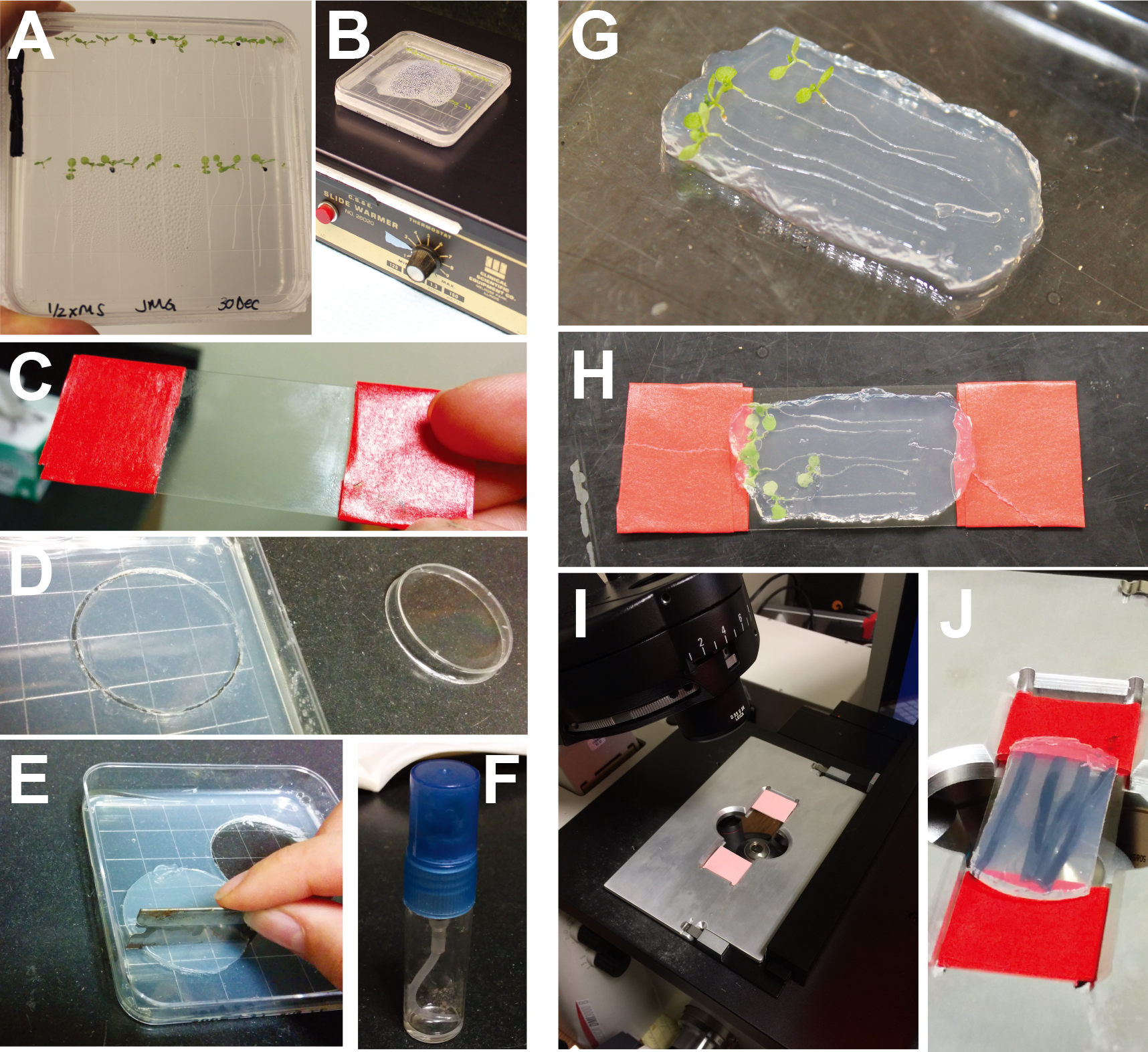
Figure 1. Experimental set-up and analysis for heat-shock degradation assay. A. 7-day old seedlings sown on square, vertically-grown 0.5x LS + 0.8% agar plates. B. Seedling plate undergoing heat shock on a slide warmer. C. Cover slip wrapped with lab tape to fit in a slide holder. D. Disc cut from coverslip media agar using a small Petri dish. E. Trimming edges off of disc so that it will fit evenly on the coverslip. F. Perfume bottle containing 0.5x LS media with ethanol or 5 μM IAA. G. Agar pad with seedlings arranged on top. H. Cover slip placed on top of agar pad and flipped over. I. Coverslip placed in the microscope slide holder. J. Close-up image of agar-plant-coverslip in slide holder.
- After 7 days of growth, place plates horizontally on a slide warmer set at 37 °C for 2 h to induce expression of the VENUS-IAA protein (Figures 1B and 2A).
- Auxin-induced VENUS-Aux/IAA degradation assay
- Prepare 0.5x LS Media + 1.2% agar coverslip media. This agar concentration is critical because it is rigid enough to handle, but low enough concentration to reduce background autofluorescence during fluorescence imaging.
- Pour 4 plates in a sterile hood, adding IAA to 2 plates and ethanol (mock) to 2 plates. To do this, pour 50 ml of media into a sterile 50 ml tube, add 50 μl of 5 mM IAA or 95% ethanol. Mix by gentle inversions and pour into two square plates.
- Prepare a rectangular 24 x 50 mm coverslip to fit within a microscope slide holder. Wrap lab tape
around each end ~3 times in order to add length so that the coverslip will fit snugly in the holder (Figure 1C). These thin coverslips break easily, so be sure to handle gently.
- When coverslip media plates are solid, use a 35 mm round Petri dish to cut circles into the agar (Figure 1D). Use a razor blade to further cut the circle so that the agar slice will fit over the coverslip and plants (Figure 1E). While cutting a circle with the small Petri dish is not a necessity, we found the agar slices cut this way were easier to remove from the plate. Alternatively, use a razor blade to cut your slice and carefully lift it from the plate.
- Prepare 5 μM IAA or mock treatments to be sprayed onto seedlings. Pipet 4 ml liquid 0.5x LS
into two clean 5 ml perfume bottles (Figure 1F). Add 4 μl of either 95% ethanol (mock) or 5 mM IAA stock solution into the perfume bottle. Keep perfume bottles clean by washing with ethanol, and then rinsing with water and LS. Save separate, labeled bottles for mock and IAA solutions.
- Using forceps, transfer 5-6 seedlings from the plate warmer to a separate agar plate (use an extra non-supplemented plant-growth plate). Spray with liquid 0.5x LS containing either 5 μM IAA or vehicle (95% ethanol). Use enough liquid to fully cover the root tips.
- Using forceps, quickly arrange these seedlings onto an agar block (prepared in step D4) containing appropriate
treatment (mock or 5 μM IAA) (Figure 1G). Place a coverslip over the agar block sandwiching the seedlings (Figure 1H). Be careful to avoid bubbles near the root tips. Flip the agar-plant-coverslip over and place in slide holder to image (Figure 1I-J).
- Image plant root tips at 0, 10, 20, 30, 40, and 60 min post-treatment (Figure 2B). It is important to minimize time between sample prep and imaging, as well as to maintain consistency between samples. We used a Leica DMI 3000B microscope fitted with a Leica long working 40x HCX PL FLUOTAR objective (see Note 2), illuminated with a Lumencor SOLA light source, and YFP filter cube. Images were captured using Leica LAS AF version 2.6.0 software and a Leica DFC345 FX camera. It is important to image all plants with the same intensity and gain settings. We used a 1 sec exposure and 3.6 gain. Based on the microscope system and particular fluorophore, it may be necessary for each lab to determine ideal exposure and gain settings. It is important to note that photobleaching may occur. In our experiments, we used controls lines with VENUS alone (i.e., with no fused IAA protein) and also mock-treated VENUS-IAA lines to test and document the extent of photobleaching that occurs over the imaging time course (Guseman et al., 2015; Moss et al., 2015).
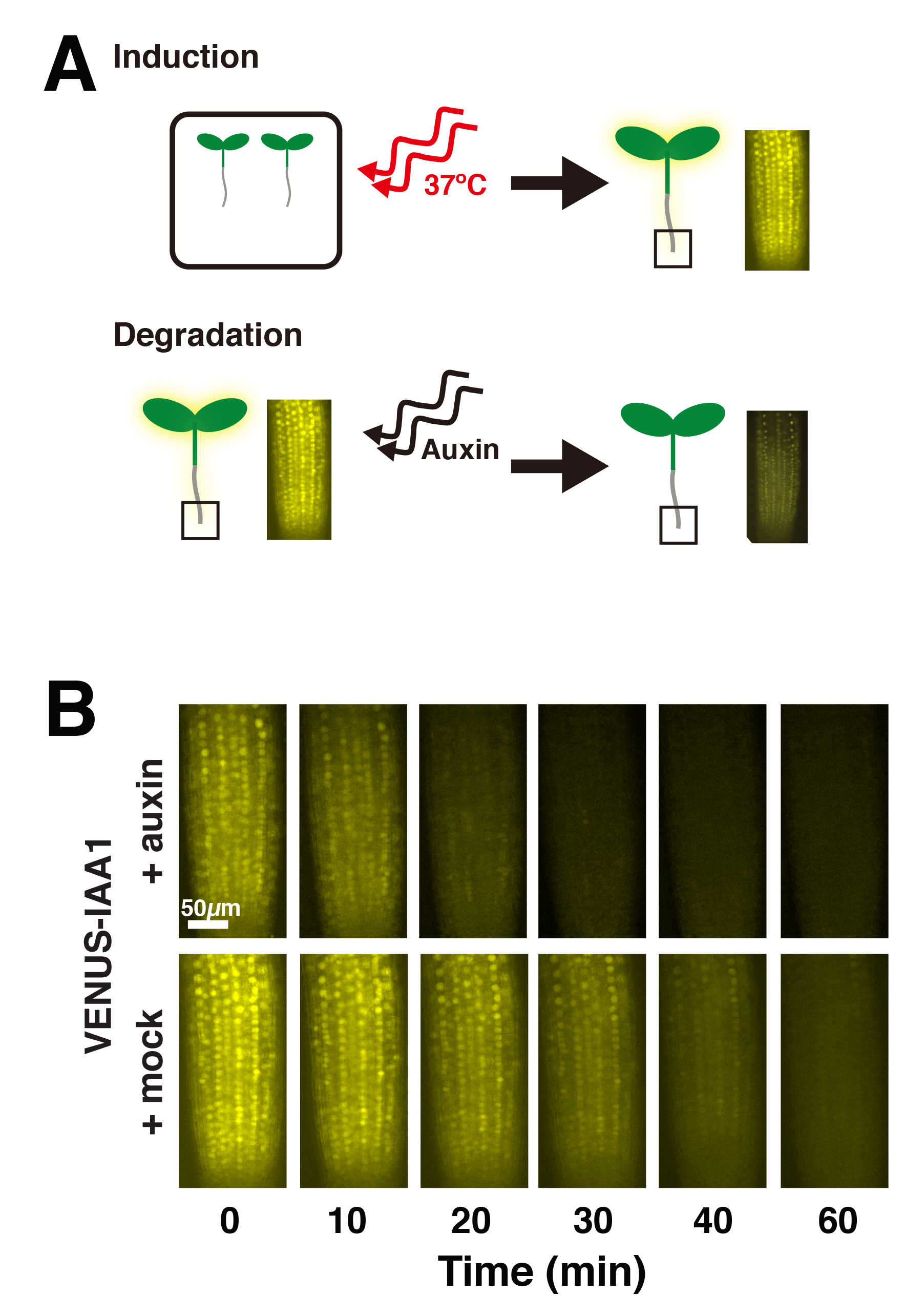
Figure 2. Schematic of auxin-induced VENUS-Aux/IAA degradation (A) and representative fluorescent root tip images following auxin or mock treatment (B)
- Prepare 0.5x LS Media + 1.2% agar coverslip media. This agar concentration is critical because it is rigid enough to handle, but low enough concentration to reduce background autofluorescence during fluorescence imaging.
- Fluorescence quantification and analysis
- Using Fiji software, open image files making sure that “autoscale” is not checked at the bottom left of the initial pop-up window. Images will all
initially appear black. The images for individual plants should be arranged into a stack of time points (Figure 3A-B). Set all images to the same brightness/contrast by using the Image > Adjust > Brightness/Contrast function for a single image, and then check the “Propagate to all other open images” box. We used a minimum and maximum of 500 and 2,200, respectively (Figure 3C).
- Using the box tool, create a box the size of the root tip region of interest (Figure 3D). Rotate the box to fit by using the Edit > Selection > Rotate function, and then move the box over the region of interest (Figure 3D). Measure pixel intensity by pressing the “M” button. Be sure that “Mean gray value” is checked under Analyze > Set Measurements. Save the region of interest box by opening Analyze > Tools > ROI Manager, and repeat measurement with this identical box on all other images in the stack. For the next stack, you may need to rotate the box to fit the new root angle, and you can save this new angled box in the ROI manager as well.
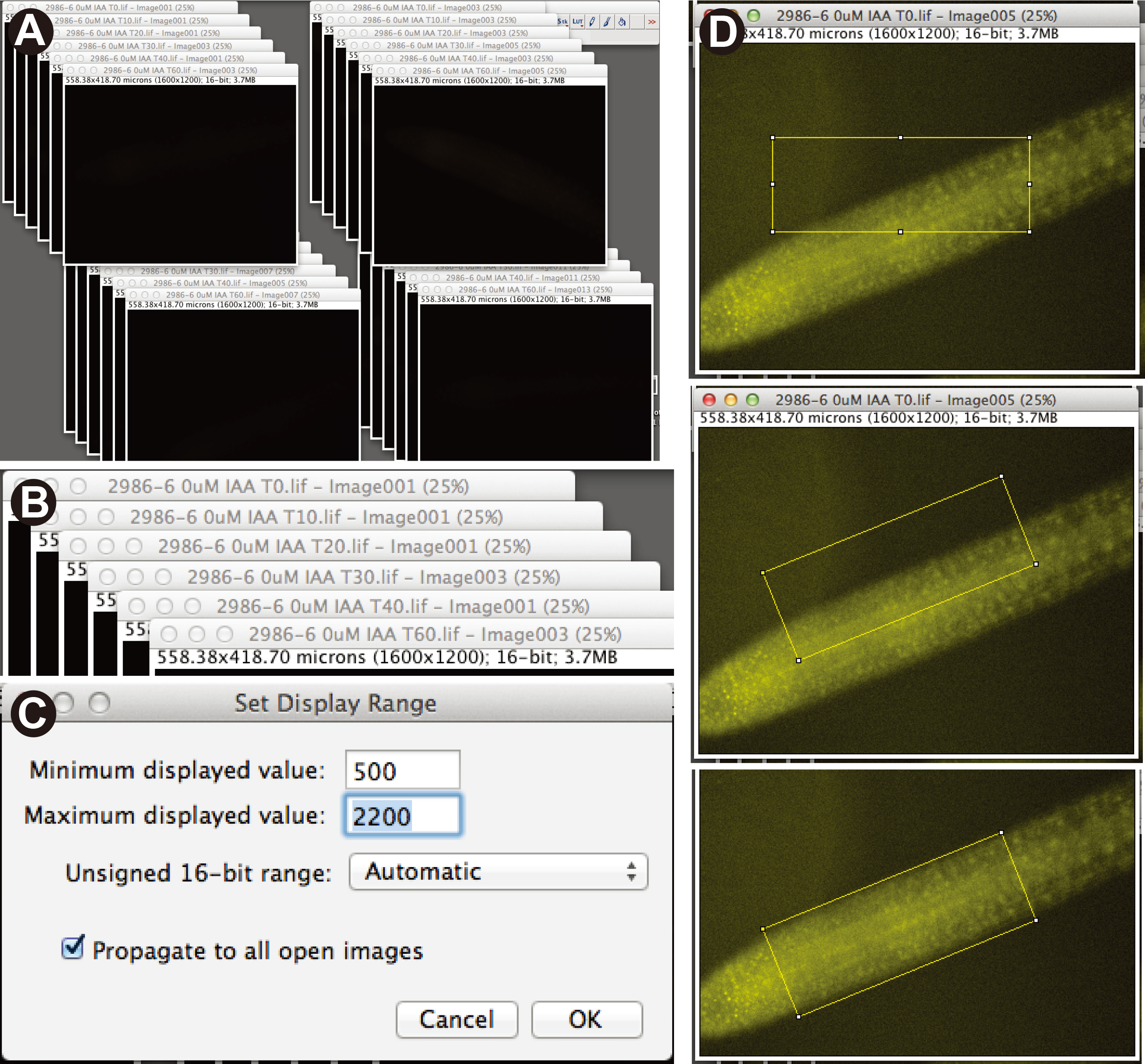
Figure 3. Analysis of heat-shock degradation assay using Fiji software. A. Image file stacks representing each individual plant from a single experiment. B. Each stack contains 6 images representing each of the time points. C. Dialog box showing Brightness/Contrast settings in Fiji. D. ROI box drawn to the width and length of the lateral root image, ROI box rotated to fit over fluorescent portion of root image, and rotated ROI box positioned of the root tip area to be measured. - Use non-fluorescent siblings (or other non-fluorescent line) to calculate background levels.
- Copy pixel intensities from the Measurements box and paste into a spreadsheet file. Label each
line, treatment, and time point. This is the raw data.
- For each line, compute the average of the background fluorescence from all non-fluorescent plants [there will likely be 2-3 (see Note 3)] at each time point. In a new column, subtract this background fluorescence value from the time point matched fluorescence value of each experimental plant (i.e., the averaged background fluorescence for time point T0 will be subtracted from all T0 values for fluorescent plants). Individual plants with T0 values lower than 350 AU were found to be within the range of noise and were removed from the dataset. Plotting the averaged background-subtracted data for each line and replicate (Figure 4A, left panel) illustrates the variability in basal fluorescence observed between lines and replicates.
- Next, normalize the fluorescence of each time point within each stack/plant by dividing by the initial fluorescence value (i.e., fold-initial normalization). Plotting the averaged normalized data for each line and replicate enables better comparison of lines and replicates (Figure 4A, right). A small number of lines behaved significantly differently from others with the same genotype (e.g., fluorescence was much higher and did not respond to auxin treatment), likely due to position effect or faulty integration of the T-DNA during transformation. These lines were removed from the analysis.
- Finally, plot the fold-initial fluorescence values averaged for all individuals from all lines and replicates. Example graphs (Figure 4B) show differences in auxin-induced degradation dynamics between VENUS-IAA1 and VENUS-IAA28.
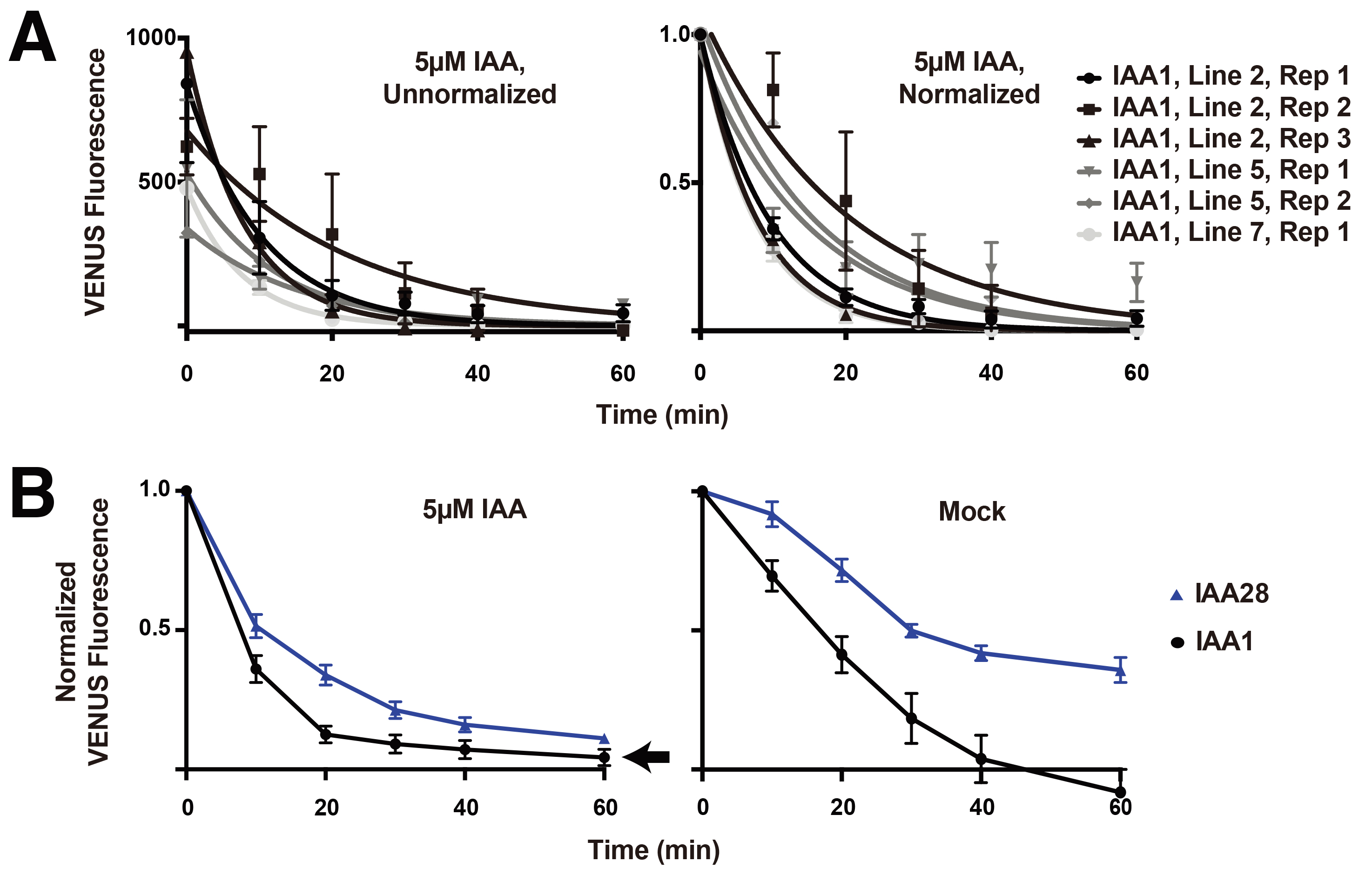
Figure 4. Representative quantification of VENUS-Aux/IAA degradation following auxin or mock treatment. A. Background-subtracted fluorescence measurements for three independent VENUS-IAA1 lines. The data is not normalized in the left panel, and is fold-initial normalized in the right panel. Each data point represents mean fluorescence from 5-6 plants carried out over replicate experiments with error bars representing standard error of the mean. B. Fold-initial normalized data for IAA1-VENUS and IAA28- VENUS, including multiple plants from at least two replicates from 2-3 independent lines. Error bars represent standard error of the mean.
- Using Fiji software, open image files making sure that “autoscale” is not checked at the bottom left of the initial pop-up window. Images will all
initially appear black. The images for individual plants should be arranged into a stack of time points (Figure 3A-B). Set all images to the same brightness/contrast by using the Image > Adjust > Brightness/Contrast function for a single image, and then check the “Propagate to all other open images” box. We used a minimum and maximum of 500 and 2,200, respectively (Figure 3C).
Notes
- We used LS media for all of our experiments. We have not tried these experiments with MS media; however, we assume it is likely that MS can be used in place of LS.
- The microscope we used is an inverted scope with a 40x long working distance (LWD) objective. The LWD objective is not required, but a 40x objective will work best. Also, if the scope used is not inverted, the agar-plant-coverslip may be placed upright on a slide for imaging.
- We used segregating T2 HS::IAA-VENUS lines that included plants both hemi- and homozygous for the T-DNA, as well as non-fluorescing plants. Consistent with this mixed population of genotypes, we did observe some variability in absolute fluorescence intensity in our raw data. Fold-initial normalization corrected for this variability and revealed the same degradation trend within each line. We used the non-fluorescent siblings to calculate background fluorescence. If you would like to use homozygous T3 lines, you will also need to grow non-fluorescent plants in parallel and use 2-3 per experiment for background fluorescence measurements.
- To further confirm the genotype of the transgenic plants, one can isolate genomic DNA from each seedling (A small leaf or two from 2-3 week old seedlings should suffice.) and perform a genotyping PCR using primers to amplify the transgene. For example, we used a forward primer binding to the HS promoter region, and a reverse primer binding to the specific Aux/IAA sequence.
- The number of plants to test for each line will depend on the variability you see within the line. Some lines are far more variable. In general, we treated 5-6 plants per line per experiment and repeated the experiment at least twice.
Recipes
- Seed sterilization solution
35 ml of 100% ethanol
25 μl of 100% Triton X-100
Fill to 50 ml with sterile water
Invert several times to mix - 0.5x Linsmaier-Skoog (LS) liquid media
Add 1,800 ml water to a beaker with stir bar. While stirring, add 1 LS packet to water. Once dissolved, bring volume up to 2 L with water. Pour 500 ml each into a separate 1 L glass bottle and autoclave (121 °C) for 20 min.
Immediately use the remaining solution to prepare agar media (see below). - Sterile plating agar solution
0.1% Bacto-agar in sterile water
Autoclave (121 °C) for 20 min - 0.5x LS media + 0.8% agar
From the remaining media above, pour 500 ml into each of two separate 1 L glass bottles. Add 4 g agar to each bottle (= 0.8% agar) and autoclave (121 °C) for 20 min. - Cover slip media (0.5x LS Media + 1.2% agar)
Pour the remaining 500 ml of 0.5x LS liquid media into a 1 L glass bottle. Add 6 g of agar (=1.2% agar) and autoclave (121 °C) for 20 min. - IAA stock solution (5 mM)
First make a 100 mM stock solution:
8.76 mg indole-3-acetic acid (IAA, MW: 175.18)
500 μl 95% ethanol
Dilute 100 mM stock with 95% ethanol to make a 5 mM solution. These stocks should be stored in the dark at -20 °C.
Acknowledgments
This protocol was adapted from Gray et al. (2001), and it was performed by Guseman et al. (2015) and Moss et al. (2015). This work was supported by the Paul G. Allen Family Foundation (J. L. N.), National Science Foundation (MCB-1411949 to J. L. N.) and National Institute of Health (R01-GM107084 to J. L. N.). B. L. M. received fellowship support from the National Cancer Institute of the National Institutes of Health (F32CA180514). J. M. G. was supported by the Developmental Biology Predoctoral Training Grant [T32HD007183] from the National Institute of Child Health and Human Development (NICHD). The authors thank Jodi LS Lilley for careful reading of this manuscript.
References
- Clough, S. J. and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6): 735-743.
- Gray, W. M., Kepinski, S., Rouse, D., Leyser, O. and Estelle, M. (2001). Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414(6861): 271-276.
- Guseman, J. M., Hellmuth, A., Lanctot, A., Feldman, T. P., Moss, B. L., Klavins, E., Calderon Villalobos, L. I. and Nemhauser, J. L. (2015). Auxin-induced degradation dynamics set the pace for lateral root development. Development 142(5): 905-909.
- Havens, K. A., Guseman, J. M., Jang, S. S., Pierre-Jerome, E., Bolten, N., Klavins, E. and Nemhauser, J. L. (2012). A synthetic approach reveals extensive tunability of auxin signaling. Plant Physiol 160(1): 135-142.
- Korasick, D. A., Jez, J. M. and Strader, L. C. (2015). Refining the nuclear auxin response pathway through structural biology. Curr Opin Plant Biol 27: 22-28.
- Moss, B. L., Mao, H., Guseman, J. M., Hinds, T. R., Hellmuth, A., Kovenock, M., Noorassa, A., Lanctot, A., Villalobos, L. I., Zheng, N. and Nemhauser, J. L. (2015). Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol 169(1): 803-813.
- Purvis, J. E. and Lahav, G. (2013). Encoding and decoding cellular information through signaling dynamics. Cell 152(5): 945-956.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Guseman, J. M., Nemhauser, J. L. and Moss, B. L. (2016). A Live-imaging, Heat Shock-inducible System to Measure Aux/IAA Degradation Rates in Planta. Bio-protocol 6(15): e1881. DOI: 10.21769/BioProtoc.1881.
- Moss, B. L., Mao, H., Guseman, J. M., Hinds, T. R., Hellmuth, A., Kovenock, M., Noorassa, A., Lanctot, A., Villalobos, L. I., Zheng, N. and Nemhauser, J. L. (2015). Rate motifs tune auxin/indole-3-acetic acid degradation dynamics. Plant Physiol 169(1): 803-813.
Category
Cell Biology > Cell imaging > Live-cell imaging
Plant Science > Plant biochemistry > Plant hormone
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









