- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Subchromoplast Fractionation Protocol for Different Solanaceae Fruit Species
Published: Vol 6, Iss 13, Jul 5, 2016 DOI: 10.21769/BioProtoc.1861 Views: 9878
Reviewed by: Ru ZhangArsalan Daudi
Abstract
Macromolecules, proteins, lipids, and other small molecules, such as carotenoids can be studied within different tissues and organelles using an array of in vitro and in vivo methodologies. In the case of tomato and other fleshy fruit the predominant organelle in ripe fruit is the chromoplast. The characteristic feature of this organelle is the presence of pigments, carotenoids at high levels. In order to fully understand the underlying biological mechanisms that operate within the chromoplast, it is necessary to perform studies at the subchromoplast level. This protocol allows the separation of plastoglobules (lipoprotein particles, which are coupled to thylakoid membranes in the chloroplasts) and membranes (thylakoid, envelope-like) of chromoplasts through a sucrose gradient. The subchromoplast compartments can then be analysed independently. Comparisons between mutant/transgenic genotypes and their backgrounds can be performed accurately with simultaneous processing during the same fractionation run. The procedure was initially developed for ripening tomato fruit but translation to sweet and hot pepper has been shown.
Materials and Reagents
- 50 ml Falcon tubes (Sigma-Aldrich, catalog number: Greiner227261 )
- Muslin (MacCulloch & Wallis, catalog number: 4470 )
- Plants
- Tomato (90 to 150 g of breaker +3 to 5 days fruit per condition)
- Sweet bell pepper (30 to 120 g of ripe fruit per condition; depending on fruit pigment content)
- Hot chilli pepper (~30 g of ripe fruit per condition)
- Tomato (90 to 150 g of breaker +3 to 5 days fruit per condition)
- DL-dithiothreitol (DTT) (Sigma-Aldrich, catalog number: 43815 )
- Sucrose (Sigma-Aldrich, catalog number: S0389 )
- Trizma base (Tris) (Sigma-Aldrich, catalog number: T1503 )
- EDTA (Sigma-Aldrich, catalog number: E5134 )
- Tricine (Sigma-Aldrich, catalog number: T0377 )
- Sodium bisulphite (sodium metabisulfite) (Sigma-Aldrich, catalog number: S9000 )
- Extraction buffer (see Recipes)
- Gradient buffers (see Recipes)
Equipment
- Cold room (4 °C)
- -20 °C room
- Centrifuge Sorvall RC-5C (Thermo Scientific)
- Fixed angle rotor Sorvall GSA-3 (purple colour) (Thermo Scientific)
- Fixed angle rotor Sorvall GSA-5 (green colour) (Thermo Scientific)
- Ultracentrifuge Beckman L7 (Beckman Coulter)
- Swing rotor SW28 (Beckman Coulter)
- 500 ml centrifuge bottles (Thermo Fisher Scientific, NalgeneTM, model: 3141-0500 )
- 50 ml centrifuge tubes (Thick-wall UltraTubes) (Thermo Fisher Scientific, NalgeneTM, model: 3110-0500 )
- 38.5 ml centrifuge tubes (Beckman Coulter, Ultra-ClearTM, model: 344058 )
- Glass flask conical narrow neck 5 L (Fischer Scientific, FisherbrandTM, model: 11597422 )
Note: This product has been withdrawn by the manufacturer. [Replacement item: Borosilicate glass narrow neck Erlenmeyer flasks (Fischer Scientific, FisherbrandTM, model: 15479103 )] - Waring blender (small laboratory blender 8010ES) (Scientific Labs, model: MIX1126 )
- Potter-Elvehjem tissue grinder (VWR International, model: 14231-372 )
- Fraction collector (Gilson, model: 203B )
- Suitable ice containers
- Funnel (glass or plastic, to fit 5 L conical flasks)
Procedure
- Rapid homogenisation and isolation of the crude chromoplast fraction
Day 0:- Harvest 90 to 150 g of tomatoes at the ripening stage of breaker + 3 to + 5 days per condition (for instance, wild type and transgenic tomatoes). The fruits need to be firm. Ensure an equal weight is used for both conditions (WT and transgenic tomatoes). For pepper fruits, harvest 30 to 120 g of fruit depending on the type of pepper and the colour of the fruit. This step varies enormously depending on the fruit, whether they contain high pigment (carotenoids) levels or not. Care must be taken at this step to avoid the use of too much fruit, as this will result in overloaded sucrose gradients and consequently, a clear separation will not be achieved. On the contrary, if there is not enough fruit, no enriched material within the gradient will be observed. It is advisable to perform several trials to optimise the correct amount of fruit needed for your experiment and laboratory conditions.
Note: Red bell peppers contain higher levels of pigments compared to orange or yellow bell peppers. So, we recommend to use 30 g of red bell peppers and 120 g of orange or yellow bell peppers. We also use 30 g for red chili peppers. However, if you are studying a high pigment or low pigment line, these weights might need to be optimised. It is only by doing the fractionation that you will see if your gradient is overloaded (same colour throughout, no clear separation). If it is the case, you will have to use less fruits. On the contrary, if you don’t see a lot of colour or no crystal, that may indicate that you did not use enough fruits to start with and you need to increase the initial fruit weight. - Wash the fruit with tap water or distilled water, deseed them.
- Cut the pericarp into pieces (± 1 cm2) (Figure 1).

Figure 1. Examples of Bell and Chilli pepper material prepared ready for homogenisation. A, B, C, and 1, 2, and 3 indicate the different accessions of pepper studied. - Place them in a plastic tray, covered with foil, overnight at 4 °C (in order to reduce the amount of starch in the fruit). This is particularly important when carrying out the procedure with green chloroplast containing fruit. It is also important to note that some mutants have altered ripening and thus carbohydrate levels and texture will vary.
- Prepare the extraction buffer (2 L) and the gradient buffers (without the DTT) and place them overnight in the cold room.
- In the -20 °C room, store the plastic containers (for the ice), eight centrifuge tubes (50 ml), four centrifuge bottles (500 ml), and two glass flasks (5 L). The plastic ware can be pre-washed with distilled water.
- Place the centrifuge rotors in the fridge overnight.
Day 1: - Start promptly in the morning (8.00 am), switch on the centrifuges and cool them to 4 °C.
- Place the rotor GSA-3 in the centrifuge RC-5C.
- Place the glass flasks and the centrifuge bottles (500 ml) on ice (in the ice containers) and place the experimental materials in the cold room.
Note: Work in the cold room in the following steps (steps 4-19 of Day 1), with everything on ice where possible. - Place the pieces of fruits in the blender (Clean the blender vessel with distilled water before re-use it for the next condition).
- Add extraction buffer (do not forget to add the DTT from the stock solution) to cover the fruits (1:3 ratio, the fruit pieces need to be adequately covered) (Figure 2).

Figure 2. Pepper pericarp in extraction buffer before homogenisation - Leave the buffer to infiltrate into the tissues for 5 min.
- Use rapid blast for 3 sec (5 sec for pepper fruit) from the Waring blender, repeated twice at high speed. This should break the cell walls but keep the chromoplasts intact, due to the sucrose maintaining a constant osmotic pressure (Figure 3).
Note: It is important that the tissues are not over homogenised, as this will result in broken plastids and the formation of gelatinous carbohydrate particulate material.
Figure 3. The extract produced following rapid homogenisation and prior to filtration - Use a glass flask, a funnel and muslin to filter the slurry through 2 to 4 layers of muslin. Pass the slurry through the muslin by squeezing gently (Figure 4).
Note: It is important not to apply too much force to avoid the extrusion of polysaccharide material that will affect the pelleting of the plastid material.
Figure 4. Representation of the extract being filtered through two to four layers of muslin to create an extract free of debris - Divide the tomato juice (filtered extract) into two centrifuge bottles (500 ml). Add extraction buffer to the suspension until it reaches 2/3 of the bottle. As a result, two 500 ml centrifuge bottles per condition are generated (Figure 5).
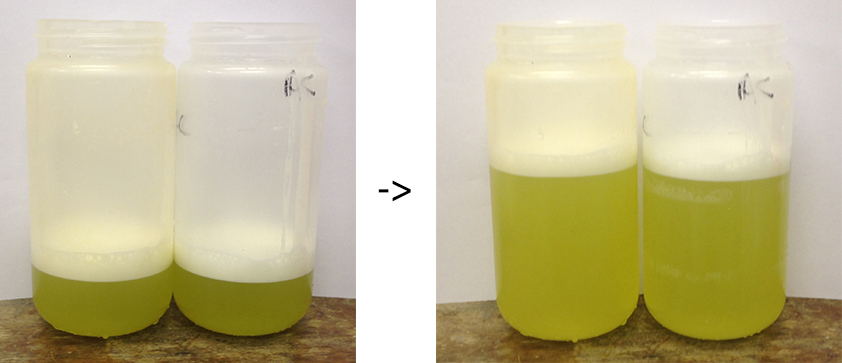
Figure 5. Examples of the extracts ready for centrifugation in suitable centrifuge pots capable of pelleting the crude plastid fractionVideo 1. Obtaining the filtered pepper extract (protocol Day 1, step 7 to 9) - Balance the centrifuge bottles carefully using a top loading balance. Tare the first one, then the second should be equivalent to ± 1 g.
- Spin the centrifuge bottles in the centrifuge (RC-5C) with the GSA-3 rotor (purple, pre-cooled) at 5,000 x g for 10 min at 4 °C. This step is to remove the cell debris (supernatant) (Figure 6).
Note: To operate the centrifuge RC-5C, check that the rotor is well attached, and close the lid by first turning the bigger cog and then the smaller one. To open it, first turn the small cog and then the big one.
Figure 6. Pepper extracts before centrifugation - Discard the supernatant carefully by pouring it from the opposite side of the pellet.
- Leave some volume (5 ml) of supernatant in the pot in order to resuspend the pellet (Figure 7).

Figure 7. Pellets representing the crude plastid fractions obtained from the pepper extracts after centrifugation - Resuspend the pellet by swirling the pots or using a glass rod with a rubber teat attached, alternatively for strong residue aggregates a pipette with a cut end tip can be used.
- Transfer the content of the centrifuge bottle in two centrifuge tubes (50 ml; this step can vary depending on the thickness and constituency of the pellet. The content of one centrifuge bottle could be transferred in only one centrifuge tube, if needed.) and then add extraction buffer in order to fill up to ¾ of the volume of the tube. At this point there are four centrifuge tubes (50 ml) per condition (Figure 8).
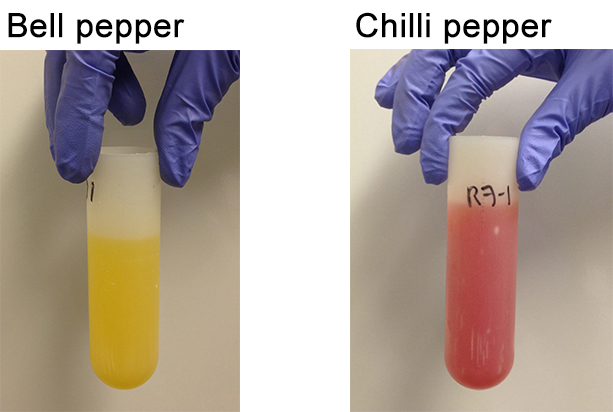
Figure 8. Pepper pellets resuspended in extraction buffer ready for the second centrifugation - Balance the centrifuge tubes carefully.
- Spin the tubes with the centrifuge RC-5C using the GSA-5 rotor (green) at 9, 000 x g for 10 min at 4 °C to pellet the plastid (Potentially, there can be some nuclear and mitochondrial contamination at this point.).
- Discard the supernatant carefully by pouring it from the opposite side of the pellet.
- This time, discard all the supernatant. The pellet should be “silky” packed in a manner that does not allow slippage (Figure 9).
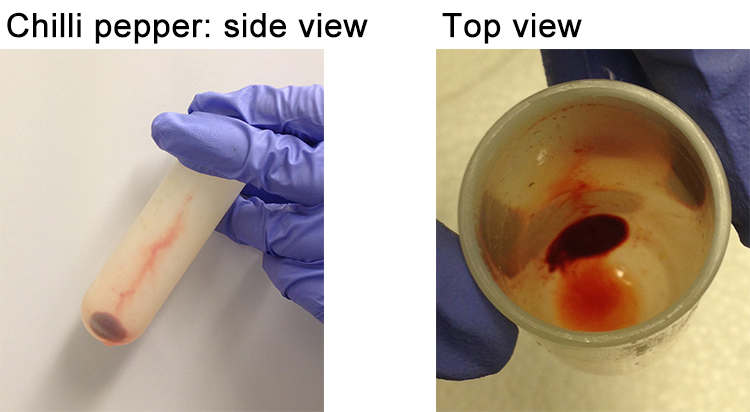
Figure 9. Pepper pellet obtained after the second centrifugation
- Harvest 90 to 150 g of tomatoes at the ripening stage of breaker + 3 to + 5 days per condition (for instance, wild type and transgenic tomatoes). The fruits need to be firm. Ensure an equal weight is used for both conditions (WT and transgenic tomatoes). For pepper fruits, harvest 30 to 120 g of fruit depending on the type of pepper and the colour of the fruit. This step varies enormously depending on the fruit, whether they contain high pigment (carotenoids) levels or not. Care must be taken at this step to avoid the use of too much fruit, as this will result in overloaded sucrose gradients and consequently, a clear separation will not be achieved. On the contrary, if there is not enough fruit, no enriched material within the gradient will be observed. It is advisable to perform several trials to optimise the correct amount of fruit needed for your experiment and laboratory conditions.
- Breaking the chromoplasts
- Resuspend the pellet of each tube with 3 ml of the 45% sucrose gradient buffer. The tubes can be vortexed. Pour and pool the chromoplast juice from the 50 ml centrifuge tubes of the same condition into the potter-Elvehjem tissue grinder, which is on ice. Wash the tubes with 0.5 ml (or more if needed) of 45% sucrose buffer and pour it into the potter-Elvehjem as well.
- Homogenise the chromoplast juice using the potter-Elvehjem (approximately 10 times) to break the chromoplasts (Video 2). Keep it vertical to avoid the glass potter from breaking (Figure 10).Video 2. Homegenisation of the pepper chromoplast juice using a potter-Elvehjem tissue grinder

Figure 10. Homogenisation of the pepper chromoplast juice using a potter-Elvehjem tissue grinder
- Resuspend the pellet of each tube with 3 ml of the 45% sucrose gradient buffer. The tubes can be vortexed. Pour and pool the chromoplast juice from the 50 ml centrifuge tubes of the same condition into the potter-Elvehjem tissue grinder, which is on ice. Wash the tubes with 0.5 ml (or more if needed) of 45% sucrose buffer and pour it into the potter-Elvehjem as well.
- Separating the subchromoplast compartments using a sucrose gradient
- Pour everything (now the subchromoplast compartment suspension) into a labelled Falcon tube (50 ml). Keep on ice. To wash the potter-Elvehjem, use 5 ml of 45% buffer twice and add it into the Falcon tube.
- Prepare the sucrose gradient in three 38.5 ml Ultra-ClearTM centrifuge tubes per condition (if not enough of the suspension is available, prepare only two tubes per condition).
- First add 8 ml of the subchromoplast compartment juice (already in the 45% sucrose buffer) to the bottom of the tube
- Then 6 ml of 38% sucrose buffer
- Then 6 ml of 20% sucrose buffer
- Then 4 ml of 15% sucrose buffer
- And finally, 8 ml of 5% sucrose buffer to the top of the gradient.
- Add each layer drop by drop on the inner wall of the tube following a circular movement, a Pasteur pipette or a Gilson pipette can be used to perform this task (Video 3).Video 3. Preparation of the discontinuous sucrose gradient

Figure 11. Ultra-ClearTM centrifuge tube containing a tomato subchromoplast compartment suspension in sucrose gradient before centrifugation
- First add 8 ml of the subchromoplast compartment juice (already in the 45% sucrose buffer) to the bottom of the tube
- Place the 38.5 ml centrifuge tubes in their metal container ready for centrifugation. Balance them (including the lid of the metal tube) using 5% sucrose buffer so they weigh exactly the same amount. Use the Beckman support to transport the centrifuge tubes to the Beckman L7 ultracentrifuge.
- Place the tubes in the SW28 rotor (tubes are hanging). Check their position by slightly twisting them and pulling them vertically and then towards the exterior of the centrifuge.
- Spin at 24,000 rpm (100,000 x g) at 4 °C for 19 h. Choose hold “on”, vacuum “on” and press start. If there is a problem with the balance, the centrifuge will stop at around 600 rpm. If there is a problem with the vacuum, the centrifuge will stop at around 2,000 rpm.
Day 2: - Stop the centrifuge: Press the stop button and when the pressure is low switch off the vacuum. The centrifuge lid can then be opened.
- Be very careful whilst removing the tubes from the rotor.
- Put them on ice.
- Take pictures of the different gradients (Figure 12).
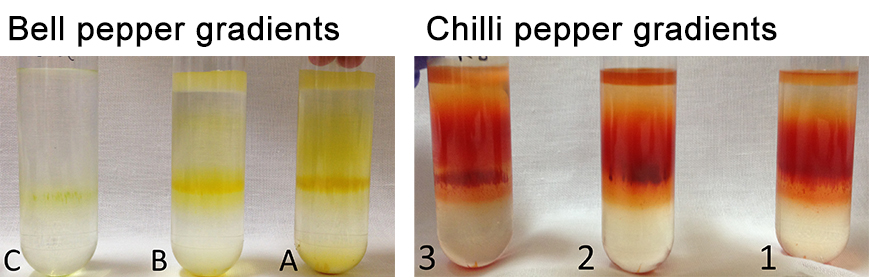
Figure 12. Separation of the pepper subchromoplast compartments in sucrose gradient for the different accessions of pepper (A, B, C, and 1, 2, 3) - Collect the fractions with the Gilson 203B fraction collector & pump system (Figure 13). Set the speed to 24.5 on the pump system (or -22.5 with anticlockwise) in order to collect 1 ml in each 1.5 ml tube. Click on the clock arrow. Keep the needle in the blank (water) for the first minute, then put it in your tube and press start on the fraction collector. Always keep the needle at the surface (just under the meniscus). Do not collect the pellet, leave some millilitres at the end (the fractions that are mixed with the pellet). When all fractions are collected, remove the needle from the tube and place in the blank (1 ml). Thus, the solution in the tubing can be retrieved. Click on the “stop” button to stop the pump system and click on the “end” button to stop the fraction collector. Collect the last millilitres with a pipette in another 1.5 ml tube.
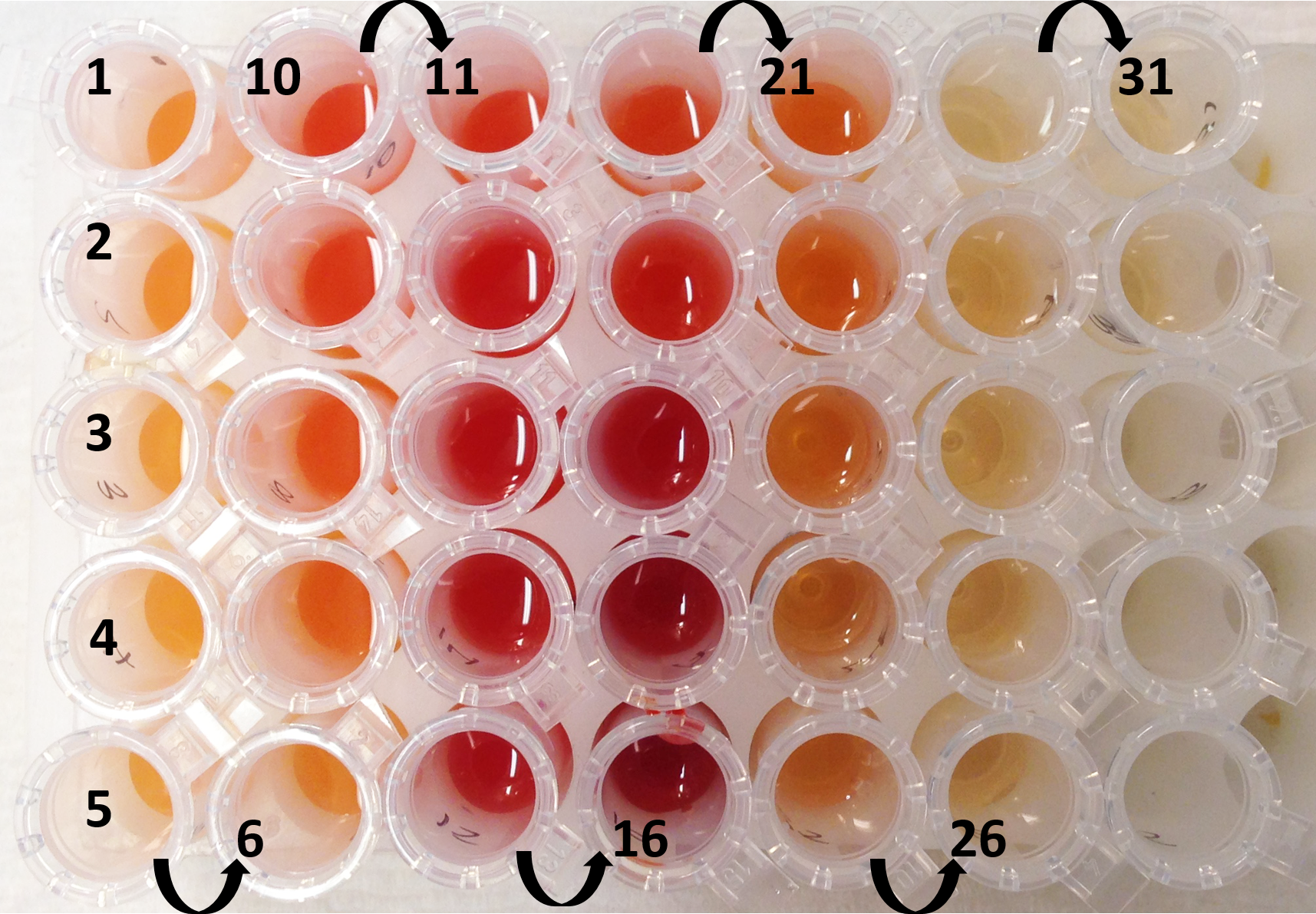
Figure 13. Chilli pepper gradient collected. Numbers indicate the order of the fractions collected (from the top of the Ultra-ClearTM centrifuge tube). - Fractions can now be analysed individually.
- Keep them at -20 °C or at -80 °C (if enzyme assays are carried out).
- Pour everything (now the subchromoplast compartment suspension) into a labelled Falcon tube (50 ml). Keep on ice. To wash the potter-Elvehjem, use 5 ml of 45% buffer twice and add it into the Falcon tube.
Representative data
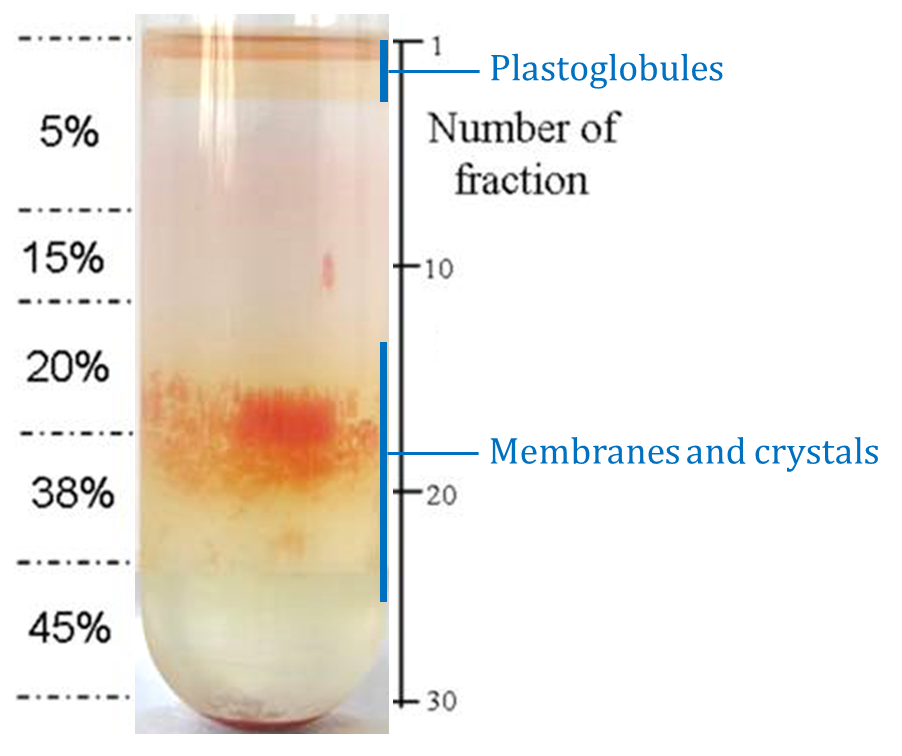
Figure 14. Subchromoplast compartments separation on the 5th step of sucrose gradient at Day 2 (Transgenic CrtB+I Ailsa Craig tomato, breaker + 3 to 5 days, 150 g). Each ml collected is called a fraction (see Day 2, point 5). The first faction corresponds to the first ml collected from the top of the tube.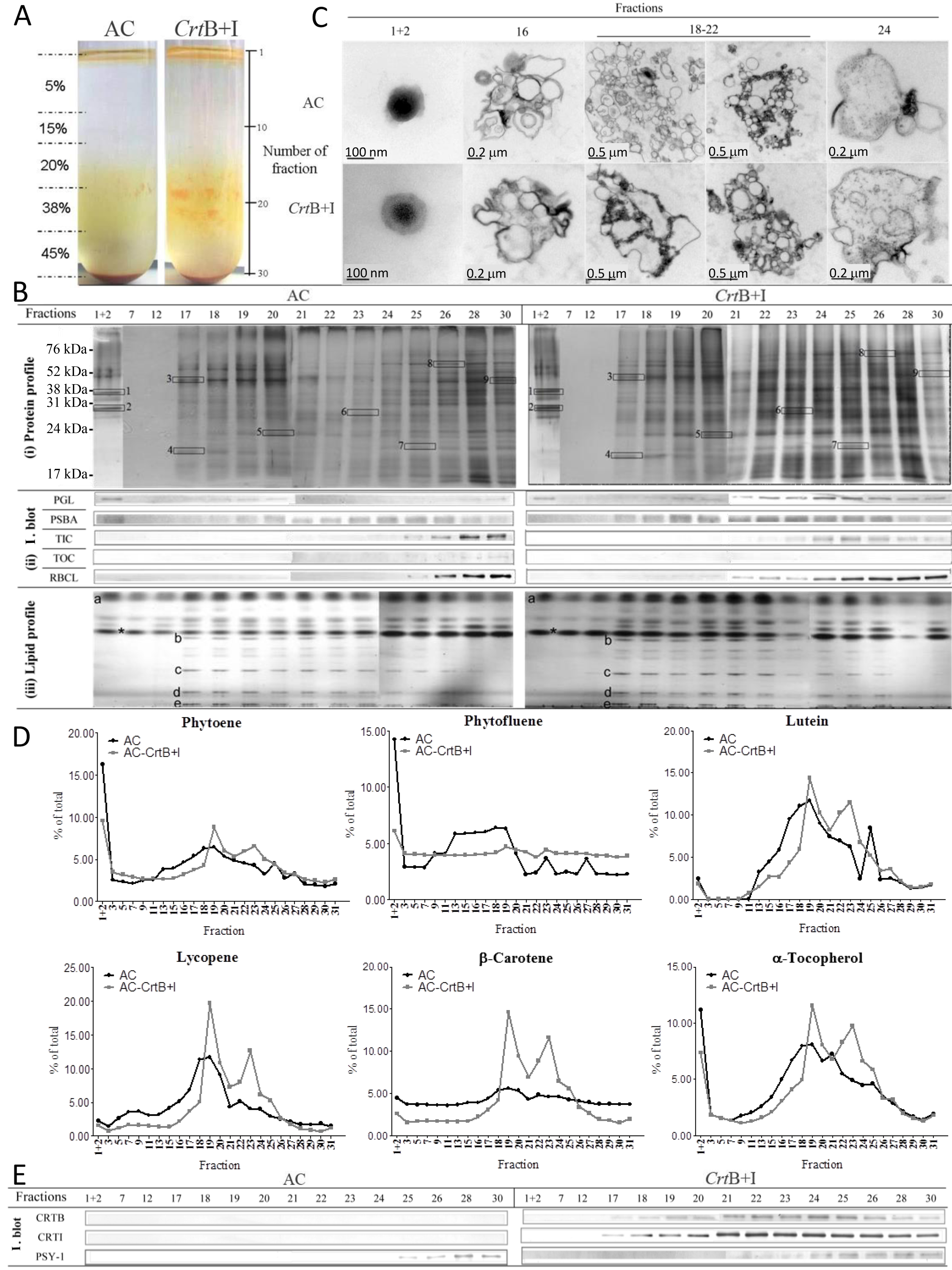
Figure 15. Example of data from Nogueira et al. (2013). A. Fractionation of subplastidial components of chromoplasts from AC (the wild type) and CrtB+I lines. Chromoplasts were extracted from 90 g of a mix of breaker + 3 to + 5 d tomatoes and then broken with a handheld potter and separated in a discontinuous gradient of 5, 15, 20, 38, and 45% sucrose (weight per volume). Fractions of 1 ml were collected for further analysis. Typically, a total of 30 fractions were collected per centrifuge tube. Fractions from six replicates were used to achieve all the experiments shown in Figure 6B. Validation of subplastidial components using antibodies to biomarker proteins and analysis of lipid species. B. (i) Protein profile. Proteins, extracted from each fraction, were separated and visualized using SDS-PAGE followed by silver staining. Selected proteins were identified by nano-LC-MS-MS: 1, Plastoglobulin-1; 2, plastid lipid-associated protein CHRC; 3, ATP synthase subunit b; 4, photosystem I reaction center subunit II; 5, photosystem II 22-kD protein; 6, oxygen evolving enhancer protein 1; 7, oxygen evolving enhancer protein 2; 8, heat shock cognate 70-kD protein-1; 9, ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit binding protein subunit b. Details of the identification of these proteins are shown in Supplemental Figure 6 online. (ii) Immunoblot (I. blot). Immunolocalization of biomarker proteins in the fractions were determined by immunoblotting: plastoglobulin (PGL, 35 kD), photosystem II protein D1 (PSBA, 28 kD), TIC (45 kD), TOC (75 kD), and ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit (RBCL, 52 kD). (iii) Lipid profile. Lipids derived from the fractions were separated in a thin layer chromatography silica plate with a mixture of acetone:toluene:water (91:30:7). Standards for lipid species were used for identification: a, triglycerides; b, monogalactodiacylglycerol; c, digalactodiacylglycerol; d, phosphatidylethanolamine; e, phosphatidylserine/phosphatidylcholine; asterisk, contaminant. C. Ultrastructure component of isolated fractions. After collection, the fractions were dialyzed against phosphate buffer and then fixed in osmium tetroxide. D. Carotenoids and a-tocopherol contents of the isolated fractions. Metabolites were extracted from each fraction and separated by liquid chromatography using a ultrahigh performance liquid chromatograph. The carotenoids and a-tocopherol were identified and quantified using calibration curves of standards. Contents are given as a percentage in a fraction compared with the total content in the tube. E. Localization of the heterologous phytoene synthase (CRTB, 38 kD) and phytoene desaturase (CRTI, 56 kD) enzymes and the endogenous phytoene synthase (PSY-1, 35 kD) enzyme within the subplastidial component of the AC and CrtB+I chromoplasts. Specific antibodies were used to immunodetect these enzymes in each collected fraction. Experiments were performed with the hemizygous CrtB+I line and a concurrent control.
Notes
The ripening stage of the fruit and the amount of fruit used are critical for this work. Here, the best combination (Breaker + 3 to 5 days & 90 to 150 g) for Ailsa Craig tomatoes is provided. However, fruits from different backgrounds can have different characteristics (firmness, quantity of pigments, etc.). Consequently, it is possible that several trials are needed before obtaining the best separation.
Recipes
- Extraction buffer
0.4 M sucrose
50 mM Trizma base
1 mM DTT
1 mM EDTA
pH = 7.8
Add the DTT (from a 1 M stock solution) just before to use the buffer - Gradient buffers
45% w/v or 38% or 20% or 15% or 5% sucrose
50 mM Tricine
2 mM EDTA
2 mM DTT
5 mM sodium bisulphite
pH = 7.9
Add the DTT (from a 1 M stock solution) just before to use the buffer
Acknowledgments
This work has been funded through the European Union’s Seventh Framework Programme for research, technological development and demonstration under grant agreement No244348 METAPRO and No613513 DISCO.
The initial preparation of the crude plastids has been reported in Fraser et al. (1994).
References
- Fraser, P. D., Truesdale, M. R., Bird, C. R., Schuch, W. and Bramley, P. M. (1994). Carotenoid biosynthesis during tomato fruit development (evidence for tissue-specific gene expression). Plant Physiol 105(1): 405-413.
- Nogueira, M., Mora, L., Enfissi, E. M., Bramley, P. M. and Fraser, P. D. (2013). Subchromoplast sequestration of carotenoids affects regulatory mechanisms in tomato lines expressing different carotenoid gene combinations. Plant Cell 25(11): 4560-4579.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Nogueira, M., Berry, H., Nohl, R., Klompmaker, M., Holden, A. and Fraser, P. D. (2016). Subchromoplast Fractionation Protocol for Different Solanaceae Fruit Species. Bio-protocol 6(13): e1861. DOI: 10.21769/BioProtoc.1861.
Category
Cell Biology > Organelle isolation > Chromoplast
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link

















