- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Protocol for Microfluidic System to Automate the Preparation and Fractionation of the Nucleic Acids in the Cytoplasm Versus Nuclei of Single Cells
(*contributed equally to this work) Published: Vol 6, Iss 12, Jun 20, 2016 DOI: 10.21769/BioProtoc.1844 Views: 9348
Reviewed by: Samik BhattacharyaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

An Optimized RNA Extraction Method From Micro-quantities of Guinea Pig Cartilage and Synovium for Osteoarthritis Research
Nidhi Bhardwaj [...] Jyotdeep Kaur
Jun 20, 2025 1617 Views

A Comparative Protocol for Preserving Deep-Water Marine Invertebrate Tissues: DNA/RNA Shield vs. Liquid Nitrogen for Dual Extraction of High-Quality Nucleic Acids
Ana S. Gomes [...] Olivier Laroche
Nov 20, 2025 1335 Views

Plasmid DNA Purification Using Filterprep With an Optional Endotoxin Removal Step
Yu-Qian Lin [...] Chung-Te Chang
Dec 20, 2025 1106 Views
Abstract
This protocol describes the extraction, fractionation, and recovery of cytoplasmic nucleic acids (e.g., cytoplasmic RNA) versus nucleic acids in the cell nucleus (including genomic DNA, gDNA) from single cells with a microfluidic system. The method enables independent, sequence-specific analyses of these critical markers (Kuriyama et al., 2015). The system uses a microfluidic chip with a simple geometry and four end-channel electrodes, and completes the entire process in less than 5 min, including lysis, purification, fractionation, and delivery to two output reservoirs: One for the nucleus (including gDNA and nuclear RNA) and one for cytoplasmic RNA. Each reservoir then contains high quality and purity aliquots with no measurable cross-contamination of cytoplasmic RNA versus nucleic acids in nucleus. As described here, our protocol focuses on the analysis of cytoplasmic RNA versus gDNA from the nucleus. We have tested this protocol with mouse and human cells but not with walled cells such as plant cells.
Keywords: Single cell analysisMaterials and Reagents
- Microfluidic chip fabrication
Fabricate a microfluidic device (Figure 1) from polydimethylsiloxane (PDMS) (Dow Corning, Sylgard® 184 Silicone Elastomer Kit) and glass slides by using soft lithography. The nominal channel width and depth of the microfluidic device are 90 μm and 35 μm, respectively. The basic design is similar to the microchannel used by Shintaku et al. (2014) except for a downstream branch channel. We have made available the CAD file for the device in the supporting information.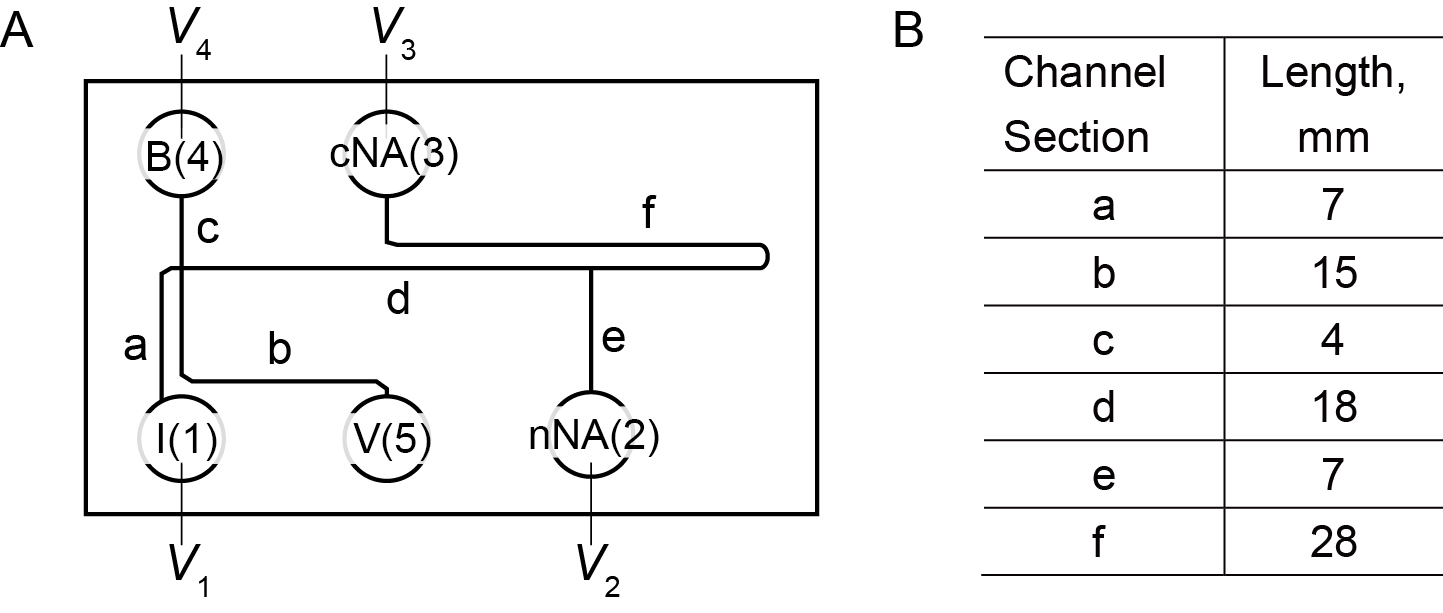
Figure 1. Schematic of channel geometry. A. The letters (a to f) in the figure identify the various channel sections. Vi (for I = 1 to 5) refer to the voltages applied to platinum electrodes placed in respective reservoirs. The numbers also refer to the reservoirs, e.g., 1 = I, 2 = nNA, 3 = cNA, 4 = B, and 5 = V. B. The lengths of channel sections in the chip. - Reagents for extraction
Prepare following solutions in UltraPure DNase-/RNase-free deionized (DI) water (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10977 ). Prepare buffer solutions under DNA/RNA free and DNase/RNase free environment (e.g., clean room conditions)- Cells (A20, mouse B cell lymphoma)
- Sodium hydroxide (NaOH) (Sigma-Aldrich, catalog number: S8045 )
- TritonTM X-100 (Sigma-Aldrich, catalog number: X100 )
- Tris (Sigma-Aldrich, catalog number: 93362 )
- Hydrochloric acid (HCl) (Sigma-Aldrich, catalog number: 258148 )
- Polyvinylpyrrolidone (PVP) (Sigma-Aldrich, catalog number: 437190 )
- HEPES (Sigma-Aldrich, catalog number: PHG0001 )
- Sucrose (Sigma-Aldrich, catalog number: S0389 )
- Leading electrolyte (LE) (see Recipes)
- Trailing electrolyte (TE) (see Recipes)
- Cell suspension buffer (see Recipes)
- Cells (A20, mouse B cell lymphoma)
- Reagents for RT-qPCR and qPCR
The reagents described here are for an experiment similar to Kuriyama et al. (2015) wherein we performed quantitation of gDNA for the nucleus and cytoplasmic RNA for the cytoplasm. However, we note the general lysing, fractionation, and preparation protocol described here is well applicable to other studies (e.g., analysis of nuclear versus cytoplasmic fractions of RNA from single cells).- TaqMan® RT-PCR mix (2x)*
- TaqMan® gene expression assay
- TaqMan® RT enzyme mix (40x)*
- TaqMan® RNA-to-CTTM 1-step kit (Thermo Fisher Scientific, Applied BiosystemsTM, catalog number: 4392653 )
- Primer set (forward primer, reverse primer) (TaqMan® probe)
- DI water
- RT-qPCR master mix for cytoplasmic RNA analysis for two reactions (see Recipes)
- qPCR master mix for gDNA analysis for one reaction (see Recipes)
*Note: TaqMan® RNA-to-CTTM 1-step kit contains both reagents.
- TaqMan® RT-PCR mix (2x)*
Equipment
- Microscope
A phase contrast microscope fitted with a CCD camera enables monitoring the lysing and cytoplasmic nucleic acid fraction focusing via isotachophoresis (ITP) [see on-line movies by Garcia-Schwarz et al. (2012) for a typical on-chip ITP experiment.]. It also helps monitor migration of nucleus and its fractionation from the cytoplasmic fraction in the ITP zone. Avoid using fluorescence-based visualization, e.g., intercalation dye, which may interfere specificity and stringency of downstream assay. - High voltage sequencer
A voltage sequencer (e.g., LabSmith, Inc., model: HVS448-3000D) automates the lysis, extraction, and fractionation via ITP.
Note: A current measurement helps monitor the ITP zone migration and fractionation of the nucleus. - Send the voltage sequence to the voltage sequencer.
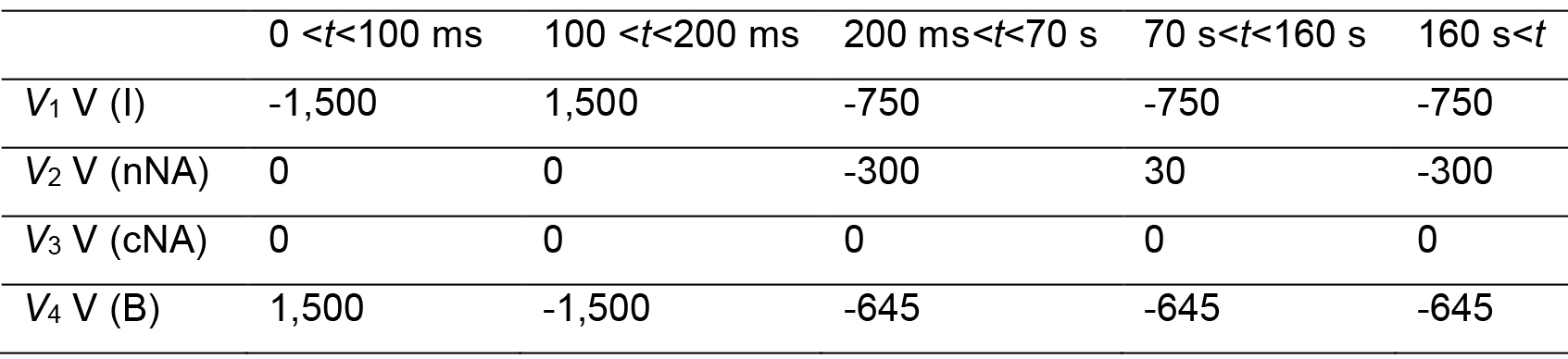
- Connect the voltage outputs to platinum wire electrodes that will be placed to the reservoirs.
Procedure
- Filling and preparation of microfluidic system
Each experiment should start with a new microfluidic system to avoid contaminating the sample. The washing protocol further decontaminates the microfluidic system and the associated platinum wire electrodes. Precondition the microchannel by filling reservoirs cytoplasmic nucleic acids out (cNA), nuclear nucleic acids out (nNA), and vacuum (V) with washing solutions (see below) and applying vacuum at reservoirs input (I) and branch (B) using a Y-connection and a single vacuum line. The washing solutions and the process are as follows (Video 1):- 1 M NaOH with 0.1% Triton X-100 for 1 min. Wash platinum wire electrodes by dipping them into the same solution.
- 1 M HCl with 0.1% Triton X-100 for 1 min. Wash platinum wire electrodes by dipping them into the same solution.
- Deionized water with 0.1% Triton X-100 for 1 min. Wash platinum wire electrodes by dipping them into the same solution.
- Dry the microchannel by applying vacuum to reservoirs I and V for 1 min.
- Load LE to reservoirs cNA, nNA and V, and apply vacuum to reservoirs I and B for approximately 1 min.Video 1. Filling and preparation of microfluidic system
- 1 M NaOH with 0.1% Triton X-100 for 1 min. Wash platinum wire electrodes by dipping them into the same solution.
- Loading single cell and extraction (Video 2)
The protocol described here ends up with 20 μl of cytoplasmic RNA sample. The protocol for 10 μl of cytoplasmic RNA sample is also available using the volumes in parentheses.- Remove the residual solution from all the reservoirs and load 20 μl (10 μl) of LE to reservoirs cNA, nNA, and V, and 20 μl (10 μl) of TE to reservoir B.
- Load a 2 μl (1 μl) cell suspension (approximately 5 cells/μl) into reservoir I. Introduce individual cells from this low concentration solution by applying a vacuum to reservoir V.
- Visually confirm an isolated single cell in the section a of the channel, between reservoir I and the cross-junction, and add 20 μl (10 μl) of the TE buffer to reservoir I.
- Place platinum wire electrodes into reservoirs I, B, cNA, and nNA, and initiate the voltage sequence for lysis, extraction and fractionation.
- Extract a nucleus from DNA Out with a 1 μl micropipette and transfer to a PCR tube containing 19 μl of the qPCR master mix.
- Extract 20 μl (10 μl) of RNA sample from RNA Out and transfer to a PCR tube.
- Mix 9 μl of RNA sample with an 11 μl of the RT-qPCR master mix.
- Perform RT-qPCR and qPCR analysis with 20 μl of total assay volume.Video 2. Loading single cell and extraction
- Remove the residual solution from all the reservoirs and load 20 μl (10 μl) of LE to reservoirs cNA, nNA, and V, and 20 μl (10 μl) of TE to reservoir B.
Recipes
- Leading electrolyte (LE) (pH 8.1)
50 mM Tris
25 mM HCl
0.4% PVP - Trailing electrolyte (TE) (pH 8.3)
50 mM Tris
25 mM HEPES
0.4% PVP - Cell suspension buffer (pH 8.3)
50 mM Tris
25 mM HEPES
225 mM sucrose
~5 cells/μl cells - RT-qPCR master mix for cytoplasmic RNA analysis for two reactions
20.0 μl TaqMan® RT-PCR mix (2x)*
2.0 μl TaqMan® gene expression assay
1.0 μl TaqMan® RT enzyme mix (40x)*
*Note: TaqMan® RNA-to-CTTM 1-step kit contains both reagents. - qPCR master mix for gDNA analysis for one reaction
10.0 μl TaqMan® RT-PCR mix (2x)
1.0 μl primer set (forward primer, reverse primer, and TaqMan® probe)*
8.0 μl DI water
*Note: Off-the-shelf primer sets can be available from TaqMan® copy number assays.
Acknowledgments
We gratefully acknowledge funding from the National Science Foundation under CBET-1159092. H. S. acknowledges funding from Japan Society for the Promotion of Science under 22686021, 26289035, and 26630052. H. S. was supported by fellowships from the John Mung Program of Kyoto University and Marubun Research Promotion Foundation, Japan.
References
- Garcia-Schwarz, G., Rogacs, A., Bahga, S. S. and Santiago, J. G. (2012). On-chip isotachophoresis for separation of ions and purification of nucleic acids. J Vis Exp (61): e3890.
- Kuriyama, K., Shintaku, H. and Santiago, J. G. (2015). Isotachophoresis for fractionation and recovery of cytoplasmic RNA and nucleus from single cells. Electrophoresis 36(14): 1658-1662.
- Shintaku, H., Nishikii, H., Marshall, L. A., Kotera, H. and Santiago, J. G. (2014). On-chip separation and analysis of RNA and DNA from single cells. Anal Chem 86(4): 1953-1957.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kuriyama, K., Shintaku, H. and Santiago, J. G. (2016). Protocol for Microfluidic System to Automate the Preparation and Fractionation of the Nucleic Acids in the Cytoplasm Versus Nuclei of Single Cells. Bio-protocol 6(12): e1844. DOI: 10.21769/BioProtoc.1844.
Category
Cell Biology > Single cell analysis > Microfluidics
Molecular Biology > DNA > DNA extraction
Molecular Biology > RNA > RNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












