- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Identification and Characterization of Bacterial Chemoreceptors Using Quantitative Capillary and Gradient Plate Chemotaxis Assays
Published: Vol 6, Iss 8, Apr 20, 2016 DOI: 10.21769/BioProtoc.1789 Views: 12859
Reviewed by: Valentine V TrotterSeda EkiciAmit Dey

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurements of Free-swimming Speed of Motile Salmonella Cells in Liquid Media
Yusuke V. Morimoto [...] Tohru Minamino
Jan 5, 2017 9617 Views

In vivo Analysis of Cyclic di-GMP Cyclase and Phosphodiesterase Activity in Escherichia coli Using a Vc2 Riboswitch-based Assay
Ying Liu [...] Ute Römling
Mar 5, 2018 7922 Views

Analysis of Gram-negative Bacteria Peptidoglycan by Ultra-performance Liquid Chromatography
Laura Alvarez [...] Felipe Cava
Oct 5, 2020 5450 Views
Abstract
Bacterial chemotaxis is a motility-based response that biases cell movement toward beneficial molecules, called attractants, and away from harmful molecules, also known as repellents. Since the species of the genus Pseudomonas are characterized by a metabolic versatility, these bacteria have developed chemotactic behaviors towards a wide range of different compounds. The specificity of a chemotactic response is determined by the chemoreceptor, which is at the beginning of the signaling cascade and which receives the signal input. The basic elements of a typical chemoreceptor are the periplasmic ligand binding domain (LBD), responsible for sensing environmental stimuli, and the cytosolic methyl-accepting (MA) domain, that interacts with other components of the cellular signaling cascade. Escherichia coli (E. coli), the traditional model in chemotaxis research, has 5 well-characterized chemoreceptors. However, genome sequence analyses have revealed that many other bacteria possess many more chemoreceptors, some of which with partially overlapping signal profiles. This high number of chemoreceptors complicates their study by the analysis of single chemoreceptor mutants. We have pursued an alternative strategy for chemoreceptor characterization which corresponds to the generation of chimeric receptors composed of the LBD of the chemoreceptor under investigation and the MA domain of an E. coli receptor (Tar). The chimer is then introduced into a chemoreceptor free mutant of E. coli and the chemotaxis of the resulting strain is entirely due to the action of this chimeric receptor. In this publication we describe the use of quantitative capillary and gradient plate assays to study Pseudomonas chemotaxis as well as E. coli strains harboring chimeric receptors.
Keywords: Chemotaxis capillary assaysMaterials and Reagents
- Materials
- SterilinTM Standard 90 mm Petri Dishes (Thermo Fisher Scientific, catalog number: 101/IRR )
- Microtest plate 96-well, F (SARSTEDT AG & Co, catalog number: 82.1581.501 )
- 1.5 ml Eppendorf tubes
- Square petri dishes (120 mm x 120 mm) with grid (Greiner Bio-One GmbH, catalog number: 688102 )
- Capillaries (Sigma-Aldrich, Drummond Microcaps®, catalog number: P1424 )
- Bulb for Pasteur pipette
- Erlenmeyer flasks 100 ml
- SterilinTM Standard 90 mm Petri Dishes (Thermo Fisher Scientific, catalog number: 101/IRR )
- Strains
- P. aeruginosa PAO1 (Stover et al., 2000)
- E. coli HD49 (Reyes-Darias et al., 2015a), chemoreceptor free strain E. coli UU1250 (Ames et al., 2002) harboring a plasmid encoding a chimeric receptor comprising the LBD of the PctC chemoreceptor of P. aeruginosa PAO1 (Taguchi et al., 1997; Rico-Jimenez et al., 2013) and the MA domain of the E. coli Tar receptor.
- P. aeruginosa PAO1 (Stover et al., 2000)
- Reagents
- HEPES sodium salt (Sigma-Aldrich, catalog number: H7006 )
- Potassium phosphate dibasic (HK2PO4) (Sigma-Aldrich, catalog number: P3786 )
- Sodium salicylate (Sigma-Aldrich, catalog number: S3007 )
- Chloramphenicol (Sigma-Aldrich, catalog number: C-0378 )
- Potassium phosphate monobasic (H2KPO4) (Sigma-Aldrich, catalog number: 60220 )
- Ammonium sulfate [(NH4)2SO4] (Merck Millipore Corporation, catalog number: 1.01217 )
- Sodium citrate tribasic dihydrate (Sigma-Aldrich, catalog number: C7254 )
- Magnesium sulfate heptahydrate (MgSO4.7H2O) (Sigma-Aldrich, catalog number: 63140 )
- Thiamine hydrochloride (Thiamine HCl) (Sigma-Aldrich, catalog number: 47858 )
- Glycerol (Scharlab, S.L., catalog number: GL0027005P )
- L-threonine (Sigma-Aldrich, Fluka, catalog number: 89179 )
- L-methionine (Sigma-Aldrich, catalog number: M9625 )
- L-leucine (Merck Millipore Corporation, catalog number: 5360 )
Note: Currently, it is “Merck Millipore Corporation, catalog number: 105360 ”. - L-histidine (Sigma-Aldrich, catalog number: 153688 )
- γ-Aminobutyric acid (GABA) (Sigma-Aldrich, catalog number: A2129 )
- Bacto-Agar (BD, DifcoTM, catalog number: 281230 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888 )
- Tryptone Broth (TB) medium (see Recipes)
- 0.9% NaCl solution (see Recipes)
- LB medium (see Recipes)
- 5x minimal A salts (see Recipes)
- 5 mg/ml Aminoacid-mix (see Recipes)
- Minimal A gradient plate medium (see Recipes)
- HEPES sodium salt (Sigma-Aldrich, catalog number: H7006 )
Equipment
- Spectrophotometer (Perkin Elmer, model: uv/vis lambda 20 )
- Incubators (30 °C and 37 °C) (Thermo Fisher Scientific, Heraeus, model: B6060 incubator)
- Orbital shaker incubator SH maxi (Controltécnica Instruments)
- Centrifuge Allegra X-22R (Beckman Coulter)
- pH meter GLP22 (HACH LANGE SPAIN, Crison)
- Bunsen burner
- Two pairs of tweezers
- Bulb dispenser (Drummond Scientific Company, catalog number: 1-000-9000 )
Procedure
In the first part of this publication we will describe the quantitative capillary assay using as an example the chemotaxis towards g-aminobutyrate (GABA). These experiments have shown that P. aeruginosa PAO1 shows GABA chemotaxis and that this response is mediated by the PctC chemoreceptor (Reyes-Darias et al., 2015a). To characterize in more detail the PctC chemoreceptor, we have produced a chimeric receptor (Figure 1), which is reported in further detail (Reyes-Darias et al., 2015a). There is now a significant body of information showing that different receptor chimera constructs are functional (Feng et al., 1997; Krikos et al., 1985; Kristich et al., 2003; Repik et al., 2000; Reyes-Darias et al., 2015a; Reyes-Darias et al., 2015b; Weerasuriya et al., 1998).
In the second part of this publication we will describe the gradient plate chemotaxis assay on the example of the chemoreceptor free strain E. coli UU1250 into which a construct encoding the PctC-Tar chimera had been introduced. This approach is a convenient means to determine the chemoeffector profile by in vivo experimentation.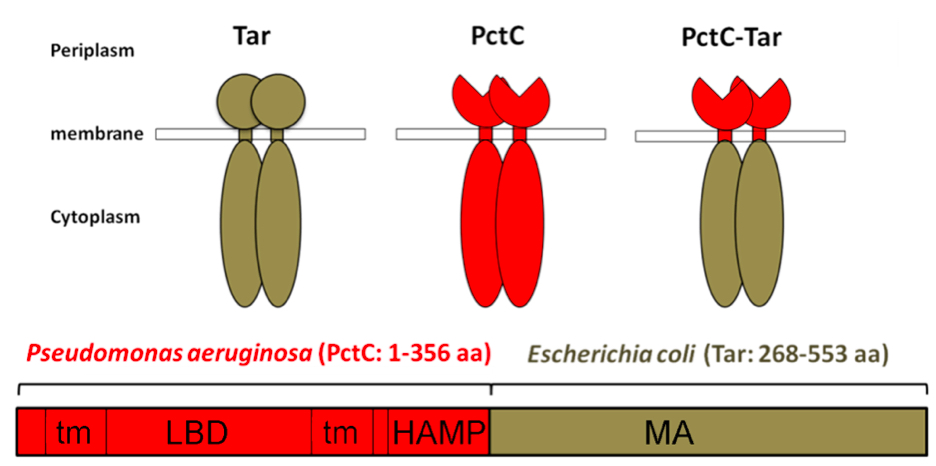
Figure 1. Construction of the PctC-Tar chimeric receptor. Tm: Transmembrane region; LBD: ligand binding domain; The HAMP linker domain (present in Histidine kinases, Adenyl cyclases, Methyl-accepting proteins and Phosphatases) is an approximately 50-amino acid alpha-helical region; MA: Methylaccepting domain; Modified version of Figure taken from (Reyes-Darias et al., 2015a).
Part I. Quantitative capillary chemotaxis assay
The principle of this assay consists of immersing chemoeffector filled capillaries into a bacterial suspension. In the case of chemotaxis, cells will preferentially swim into the capillary whereas in the case of chemorepellation, fewer cells will swim into the capillary as compared to the buffer filled control capillary. Capillaries are then emptied and the number of colony forming units is determined. We detail here a modified version of the original capillary assay developed by Adler (1973).
- P. aeruginosa PAO1 is grown overnight (12-18 h) in LB medium (pH=7.4) on a rotator shaker at 200 rpm and 37 °C.
- Subsequently 0.1-0.2 ml overnight culture is transferred to 20 ml of fresh LB medium (pH=7.4) in 100 ml Erlenmeyer flasks. The initial OD600 should be between 0.05-0.07.
- The culture is grown to early stationary phase (OD600 0.25-0.35) at 37 °C and 200 rpm.
- The cells then have to be changed to an appropriate medium for mobility and chemotaxis. Wash twice 4 ml of the culture with four ml of chemotaxis buffer (HEPES, pH=7.0) by centrifuging at 1,667 x g (4 °C) for 5 min, followed by resuspension of cell pellet to a density of OD600= 0.04-0.05 in 4-8 ml of chemotaxis buffer. Harsh cell treatment may result in the loss of the flagella.
- Subsequently, 230 μl aliquots of bacterial suspension are placed into the wells of a 96-well plate.
- Capillaries are heat-sealed at one end over the flame of a burner (Figure 2A). The open end is then inserted into the chemoattractant solution (Figure 2B), of which the pH had been adjusted to that measured for the bacterial suspension (pH=7.0). As negative controls, capillaries are filled with chemotaxis buffer and for positive controls, capillaries are filled with a known, strong chemoattractant such, in the case of Pseudomonas strains, with 0.1 % of casamino acids. The closed end of the capillary is inserted into the rubber adaptor, which is then placed onto the wells in a way that the open end of the capillary is immersed into the bacterial suspension (Figure 2C).
- After incubation for 30 min, the capillaries are removed from the plate and the section of the capillary that was in contact with bacteria is rinsed with water. The sealed end of the capillary is broken and its contents emptied into a microfuge tube containing 1 ml of 0.9% (w/v) NaCl solution (Figure 2D) using a bulb dispenser.
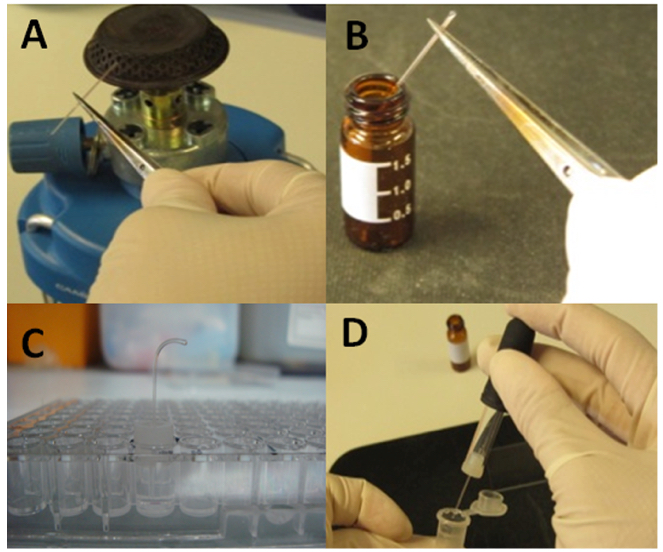
Figure 2. Experimental setup of capillary assays. A. One end of the capillary is sealed on open fire. B. Sealed and warmed capillary is inserted into the chemoattractant solution for filling. C. The rubber adaptor with the capillary is placed onto the well, which submerges the open end of capillary into the bacterial suspension that had been placed previously into the well. D. The capillary content is emptied into an Eppendorf tube containing 0.9% (w/v) NaCl.
- After a short centrifugation (5-10 sec), serial 10-fold dilutions of the resulting cell suspension are prepared in 0.9% (w/v) NaCl. For cell counting, a modified version of the protocol described in (Hoben and Somasegaran, 1982) is used. Agar plates containing LB medium (pH=7.4) supplemented with 17.5 µg/ml chloramphenicol are divided into six sectors, and three individual 20 µl aliquots are placed onto a plate sector as shown in Figure 3.
- Plates are incubated at 37 °C for 20-24 h prior to cell counting.
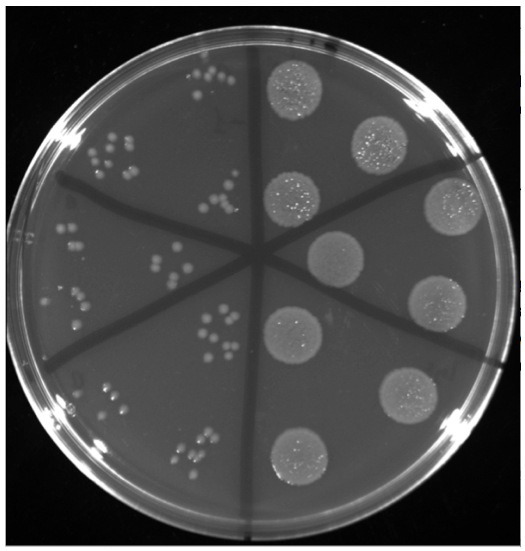
Figure 3. LB agar plates containing chloramphenicol used for cell counting in the quantitative capillary chemotaxis assay. P. aeruginosa PAO1 colonies from the buffer control (left) and from chemotaxis towards 1 mM GABA (right). Shown are 10-fold dilutions and further dilutions are necessary to precisely quantify colonies of the GABA chemotaxis experiment.
Part II. Gradient plate assays
In this assay aliquots of chemoeffectors are deposited on an agar plate. After incubation permitting chemoeffector gradient formation, aliquots of the bacterial suspension are placed at a defined distance from the deposited chemoeffector. After approx. 1 day, plates are inspected. In the case of chemoattraction, the bacterial halo is acentric towards the chemoeffector, in the case of chemorepellation the halo is acentric away from the chemoeffector.
- Square petri dishes (120 mm x 120 mm) with vents are filled with 80 ml of Minimal A gradient plate medium (pH=7) (see Note 1). Plates are cooled at room temperature for at least 3 h. Care must be taken when moving the plates since the agar is semi-solid and can be damaged easily.
- Along the vertical central line of the plate, 10 μl aliquots of a concentrated chemoeffector solution (10 mM GABA in the case shown, see Note 2), dissolved in sterile water, are placed at regular distances (Figure 4). Plates are incubated for 12-16 h at 4 °C for gradient formation.
- Bacteria, in the case shown E. coli HD49, are grown overnight in TB medium supplemented with 17.5 µg/ml chloramphenicol at 30 °C.
- 1 ml of the cell culture is then washed twice with 1 ml of 0.9 % (w/v) NaCl by consecutive centrifugation at 1,667 x g at 4 °C for 5 min and resuspension. The cell suspension is then diluted with 0.9% (w/v) NaCl to an OD600 of 0.4-0.6.
- Initially, 2 microliter aliquots of bacterial suspension are placed horizontally to each of the chemoattractant spots but with varying distances of 0.5-3.5 cm to the chemoeffector deposit (Figure 4). Plates are incubated at 30 °C for 16-30 h. The distance at which chemotaxis is seen best is then used for further analyses. Typically, cells can be deposited on either side of the attractant, which is a way to generate duplicate measurements or to assess taxis of different strains (wt and mutant strain) towards the same chemoeffector deposit.
- From these plates the magnitude of chemotaxis can be determined in a semi-quantitative manner by calculating the response index as described by (Pham and Parkinson, 2011). Briefly, the distances from the site of inoculation to the colony edges closest to (D1) and furthest from (D2) the chemoattractant source are determined to calculate the response index (RI) using RI = D1/(D1 + D2). RIs superior to 0.52 indicate chemotaxis, whereas RIs inferior to 0.48 represent chemorepellation. RI values between 0.48 and 0.52 indicate neutral behavior.
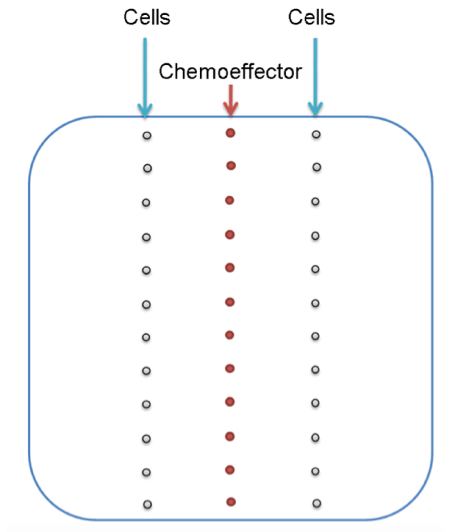
Figure 4. The experimental set-up of the gradient plate assay
Representative data
Figure 5 shows results from quantitative capillary assays of P. aeruginosa PAO1, its mutant deficient in the PctC chemoreceptor and the mutant complemented with a plasmid harboring the mcpG gene towards GABA. Data were corrected with the number of cells that swam into buffer containing capillaries and are expressed as bacterial cells that migrate into the GABA containing capillary.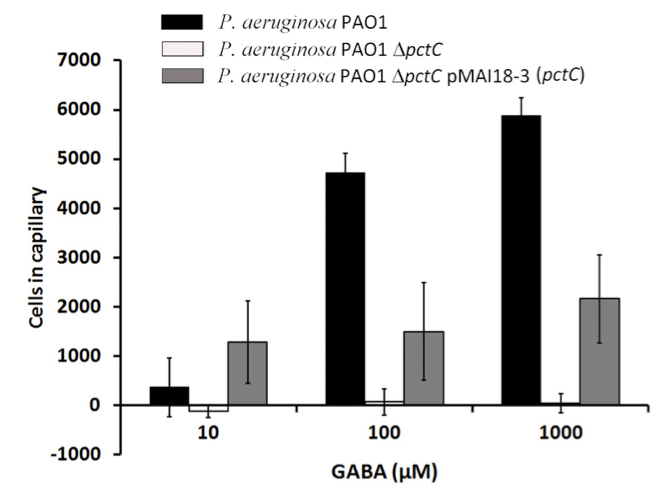
Figure 5. Quantitative capillary chemotaxis assays of wild type, mutant and complemented mutant strains of P. aeruginosa PAO1 to GABA. This figure was taken from Rico-Jimenez et al. (2013).
Figure 6 shows gradient plate assays towards GABA of E. coli strains containing either the Tar receptor or the PctC-Tar chimera as a sole chemoreceptor. In the case of Tar, the bacterial halo is circular and consequently an RI of 0.50 was determined indicating the absence of chemotaxis. In the PctC-Tar experiment, the halo is acentric towards deposited GABA and the derived RI of 0.88 indicates strong chemotaxis.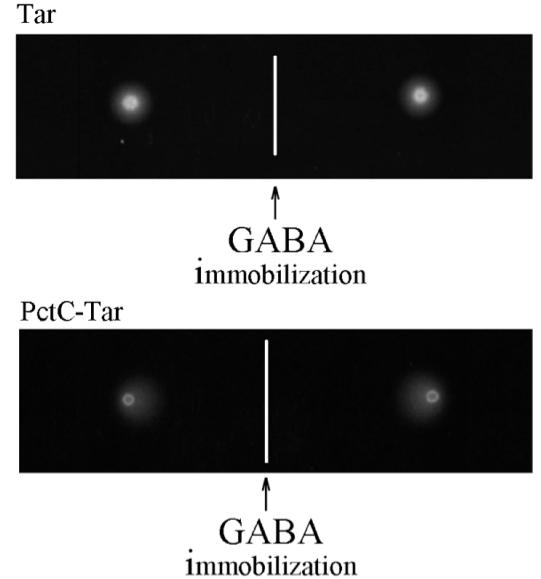
Figure 6. Gradient plate assay of E. coli expressing either Tar (top) or PctC-Tar (bottom) as the only chemoreceptor. Shown are duplicate samples. The response indices were 0.5 (± 0.05) for Tar and 0.88 (± 0.02) for PctC-Tar. Modified version of a figure is taken from Reyes-Darias et al. (2015a).
Notes
- The plate contained 1 μM sodium salicylate to induce expression of the chimeric receptors and 17.5 μg/ml chloramphenicol to select for plasmid maintenance.
- The optimal attractant concentration in the gradient plate assay varies. Initial experiments with different concentrations for chemoeffector deposit may be carried out (1 to 100 mM). Frequently, good responses are observed with 10 mM solutions and a distance of 2.5 cm between bacteria and the chemoeffector.
- Prepare all solutions using ultrapure water (prepared by purifying deionized water to attain a sensitivity of 18 MΩ cm at 25 °C) and analytical grade reagents.
Recipes
Note: Prepare all solutions using ultrapure water (prepared by purifying deionized water to attain a sensitivity of 18 MΩ cm at 25 °C) and analytical grade reagents.
- Tryptone Broth (TB) medium
10 g tryptone
5 g NaCl
Add dH2O to 1,000 ml
Autoclaved and stored at room temperature - 0.9% NaCl solution
9 g of NaCl
Add dH2O to 1,000 ml
Autoclaved and stored at room temperature - Luria-Bertani (LB) medium
5 g yeast extract
10 g tryptone broth medium
5 g NaCl
Add dH2O to 1,000 ml
Adjust pH to 7.4
Autoclaved and stored at room temperature - 5x minimal A salts
26.25 g K2HPO4
11.25 g KH2PO4
2.5 g (NH4)2SO4
1.25 g Na citrate
Add dH2O to 500 ml
Autoclaved and stored at room temperature - 5 mg/ml Aminoacid-mix
250 mg of L-threonine
250 mg of L-histidine
250 mg L-methionine
250 mg L-leucine
dH2O to 50 ml
Filtered and stored at 4 °C - Minimal A gradient plate medium
1x minimal A salts
0.2% (v/v) glycerol
1 mM MgSO4.7H2O
0.04 mg/ml aminoacid-mix
0.1 mg/ml thiamine
0.25% (w/v) agar
dH2O to 100 ml
Adjust pH to 7
Gradient plates are allowed to solidify between 2-4 h at room temperature
Acknowledgments
This work was supported by FEDER funds and Fondo Social Europeo through grants from the Junta de Andalucía (grants P09-RNM-4509 and CVI-7335 to T. K.), the Spanish Ministry for Economy and Competitiveness (grant Bio2010-16937 to T. K.) and EMBO short term fellowship grant ASTF 479 -2012 to JA-R.
References
- Adler, J. (1973). A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol 74(1): 77-91.
- Ames, P., Studdert, C. A., Reiser, R. H. and Parkinson, J. S. (2002). Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci U S A 99(10): 7060-7065.
- Feng, X., Baumgartner, J. W. and Hazelbauer, G. L. (1997). High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J Bacteriol 179(21): 6714-6720.
- Hoben, H. J. and Somasegaran, P. (1982). Comparison of the pour, spread, and drop plate methods for enumeration of rhizobium spp. in inoculants made from presterilized peat. Appl Environ Microbiol 44(5): 1246-1247.
- Krikos, A., Conley, M. P., Boyd, A., Berg, H. C. and Simon, M. I. (1985). Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci U S A 82(5): 1326-1330.
- Kristich, C. J., Glekas, G. D. and Ordal, G. W. (2003). The conserved cytoplasmic module of the transmembrane chemoreceptor McpC mediates carbohydrate chemotaxis in Bacillus subtilis. Mol Microbiol 47(5): 1353-1366.
- Pham, H. T. and Parkinson, J. S. (2011). Phenol sensing by Escherichia coli chemoreceptors: a nonclassical mechanism. J Bacteriol 193(23): 6597-6604.
- Repik, A., Rebbapragada, A., Johnson, M. S., Haznedar, J. O., Zhulin, I. B. and Taylor, B. L. (2000). PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol Microbiol 36(4): 806-816.
- Reyes-Darias, J. A., Garcia, V., Rico-Jimenez, M., Corral-Lugo, A., Lesouhaitier, O., Juarez-Hernandez, D., Yang, Y., Bi, S., Feuilloley, M., Munoz-Rojas, J., Sourjik, V. and Krell, T. (2015a). Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol Microbiol 97(3): 488-501.
- Reyes-Darias, J. A., Yang, Y., Sourjik, V. and Krell, T. (2015b). Correlation between signal input and output in PctA and PctB amino acid chemoreceptor of Pseudomonas aeruginosa. Mol Microbiol 96(3): 513-525.
- Rico-Jimenez, M., Munoz-Martinez, F., Garcia-Fontana, C., Fernandez, M., Morel, B., Ortega, A., Ramos, J. L. and Krell, T. (2013). Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol Microbiol 88(6): 1230-1243.
- Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., Brinkman, F. S., Hufnagle, W. O., Kowalik, D. J., Lagrou, M., Garber, R. L., Goltry, L., Tolentino, E., Westbrock-Wadman, S., Yuan, Y., Brody, L. L., Coulter, S. N., Folger, K. R., Kas, A., Larbig, K., Lim, R., Smith, K., Spencer, D., Wong, G. K., Wu, Z., Paulsen, I. T., Reizer, J., Saier, M. H., Hancock, R. E., Lory, S. and Olson, M. V. (2000). Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406(6799): 959-964.
- Taguchi, K., Fukutomi, H., Kuroda, A., Kato, J. and Ohtake, H. (1997). Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 143 ( Pt 10): 3223-3229.
- Weerasuriya, S., Schneider, B. M. and Manson, M. D. (1998). Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J Bacteriol 180(4): 914-920.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Reyes-Darias, J. A., García, V., Rico-Jiménez, M., Corral-Lugo, A. and Krell, T. (2016). Identification and Characterization of Bacterial Chemoreceptors Using Quantitative Capillary and Gradient Plate Chemotaxis Assays. Bio-protocol 6(8): e1789. DOI: 10.21769/BioProtoc.1789.
Category
Microbiology > Microbial cell biology > Cell-based analysis
Microbiology > Microbial signaling > Sensory receptor
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link














