- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Conjugation of Duplexed siRNN Oligonucleotides with DD-HyNic Peptides for Cellular Delivery of RNAi Triggers
Published: Vol 6, Iss 7, Apr 5, 2016 DOI: 10.21769/BioProtoc.1782 Views: 9345
Reviewed by: Arsalan DaudiVikash VermaAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Effective Gene Silencing in Plants by Synthetic Trans-Acting siRNAs Derived From Minimal Precursors
Adriana E. Cisneros [...] Alberto Carbonell
Oct 20, 2025 1813 Views

Ribozyme-Mediated Knockdown of lncRNA Gene Expression in Drosophila
Kevin G. Nyberg and Richard W. Carthew
Oct 20, 2025 2128 Views

A Highly Efficient siRNA Transfection Method in Primary Cultured Cortical Neurons
Xiaorong Wang [...] Zhaolong Zhang
Jan 20, 2026 297 Views
Abstract
Despite the great promise that short interfering RNA (siRNA) induced RNAi responses hold as a therapeutic modality, due to their size (~15 kDa) and high negative charge (Bumcrot et al., 2006), siRNAs have no bioavailability and require a delivery agent to enter cells (Figure 1). TAT peptide transduction domain (PTD) has been developed as an agent that mediates cellular delivery of macromolecular therapeutics that otherwise lack bioavailability, making it a tantalizing candidate for siRNA delivery (Farkhani et al., 2014). Unfortunately, when conjugated to TAT PTD, the presence of 40 negative phosphodiester backbone charges on siRNA neutralizes the cationic PTD resulting in aggregation and poor cellular delivery (Meade and Dowdy, 2007). In light of this, we synthesized a neutral RNAi trigger, termed siRiboNucleic Neutrals, for conjugation to TAT PTD (Meade et al., 2014). In brief, the negatively charged phosphodiester backbone was neutralized by synthesis with bio-reversible phosphotriester protecting groups which are specifically converted into charged phosphodiester bonds inside of cells by the action of cytoplasmic restricted thioesterases resulting in a wild type siRNA that can induce RNAi responses. Here we describe the conjugation and cellular delivery of siRNN oligonucleotides with TAT PTD delivery domain (DD) HyNic peptides.
Keywords: siRNAMaterials and Reagents
- 1.5 ml microcentrifuge tubes (Polypropylene, Pyrogen, RNase, DNase-free)
- Amicon Ultra 0.5 ml centrifugal filter unit with Ultracel-30 membrane (Merck Millipore Corporation, catalog number: UFC503024 )
- 50 ml conical centrifuge tubes (VWR International, catalog number: 21008-714 )
- 24-well cell culture plates, flat bottom, TC treated (Genesee Scientific, catalog number: 25-107 )
- H1299 cells (ATCC, catalog number: CRL-5803 )
Note: H1299 cells constitutively expressing destabilized enhanced green fluorescent protein (dGFP) are used in this protocol for rapid assaying of RNAi responses by flow cytometry. - GFP and non-targeting siRNNA4 (RNN phosphoramidite and oligonucleotide synthesis is described in Meade et al., 2014), RNN sequences (5’ to 3’):
- GFP passenger strand: CDCACUAACCUGAGCAACCACAGUAT
- GFP guide strand: CUSGGGUSGCUSCAGGUSAGUSGGUST
- Non-targeting passenger strand: UDGAGAAGAUCCUCAAUAAAAGAUAT
- Non-targeting guide strand: UCSUUUASUGASGGAUCSUCUSCAUST
- Key: subscript D = dimethyl-butyl phosphotriester group, subscript A = aldehyde A-SATE phosphotriester group, subscript S = tBu-SATE phosphotriester group
- GFP passenger strand: CDCACUAACCUGAGCAACCACAGUAT
- UltraPure DNase/RNase-free distilled water (Life Technologies, Invitrogen, catalog number: 10977023 )
Note: Currently, it is “Thermo Fisher Scientific, InvitrogenTM, catalog number: 10977023”. - TAT Delivery Domain (DD) Hynic peptide, 3T3S-Hy [TAT = RKKRRQRRR, 3T3S-Hy sequence: HyNic-GG-(TAT)-PEG18-(TAT)-PEG18-(TAT)] (Note 1)
- Acetonitrile (ACN), Anhydrous (Glen Research, catalog number: 40-4050-50 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S7653 )
- HEPES (AmericanBio, catalog number: AB00892 )
- Aniline (Alfa Aesar, catalog number: A14443 )
- Acetic Acid, Glacial (Thermo Fisher Scientific, catalog number: MAX00739 )
- Dry ice
- 40% acrylamide/bis solution (19:1) (Bio-Rad Laboratories, catalog number: 161-0144 )
- 10x Tris-Borate-EDTA (TBE) Buffer (Mediatech, catalog number: 46-011-CM )
- Ammonium persulfate (VWR International, catalog number: EM-2300 )
- 10% (w/v) sodium dodecyl sulfate (SDS) solution (Bio-Rad Laboratories, catalog number: 161-0416 )
- N, N, N’, N’-Tetramethylethylenediamine (TEMED) (VWR International, catalog number: EM-8920 )
- Deionized water
- Orange G (Sigma-Aldrich, catalog number: O-3756 )
- Glycerol (Sigma-Aldrich, catalog number: G5516 )
- SilverQuest Silver Staining Kit (Life Technologies, Novex™, catalog number: LC6070 )
Note: Currently, it is “Thermo Fisher Scientific, InvitrogenTM, catalog number: LC6070”. - DMEM, high glucose (Life Technologies, Gibco, catalog number: 11965-092 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 11965-092”. - Fetal Bovine Serum (FBS), heat inactivated (Omega Scientific, catalog number: FB-02 )
- Penicillin-Streptomycin (10,000 U/ml) (Life Technologies, Gibco, catalog number: 15140-122 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 15140-122”. - Phosphate buffered saline (Thermo Fisher Scientific, catalog number: BP665-1 )
- Opti-MEM reduced serum medium (Life Technologies, Gibco, catalog number: 31985-070 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 31985-070”. - 0.05% Trypsin-EDTA, phenol red (Life Technologies, Gibco, catalog number: 25300-054 )
Note: Currently, it is “Thermo Fisher Scientific, GibcoTM, catalog number: 25300-054”. - 5x siRNN conjugation buffer (see Recipes)
- 0.4% SDS non-denaturing polyacrylamide gel solution (see Recipes)
- 0.4% SDS-PAGE running buffer (see Recipes)
- 1x 0.4% SDS non-denaturing loading buffer (see Recipes)
Equipment
- Rotisserie (Barnstead International, model: Labquake 4001100 )
- Lyophilizer (SP Scientific, VirTis, model: Freezemobile 25EL )
- Microcentrifuge (Eppendorf, model: 5424 )
- UV Spectrophotometer (capable of measuring absorbance at 260 nm)
- Polyacrylamide gel casting stand and glass with 1.5 mm spacers (Bio-Rad Laboratories)
- Gel running apparatus (Bio-Rad Laboratories, model: Mini-PROTEAN 3 Cell )
- Gel staining dish
- Gel Doc (Bio-Rad Laboratories, model: Molecular Imager Gel Doc XR+ System )
- Benchtop centrifuge (Beckman Coulter, model: Allegra X-15R )
- Flow cytometer (BD Sciences, model: LSRII )
Procedure
The number of different 21-mer siRNNs that can be synthesized from only the published phosphotriester groups outnumbers the number of atoms in the universe by several orders of magnitude (Meade et al., 2014). No single siRNN configuration will be the most effective for all situations. The optimal positioning, type, and number of phosphotriester groups composing a siRNN should be determined for conjugation with a particular delivery or targeting domain and delivery into target cells or tissues. In this protocol, we focus on conjugation, purification, and cellular delivery of duplexed siRNNA4 oligonucleotides with a TAT Delivery Domain (DD) peptide as described in Meade et al. (2014) (Figure 2).
- siRNN conjugation and purification
- Set up the following conjugation reaction in a 1.5 ml microcentrifuge tube in 50% ACN /50% ultrapure water (Note 2):
- 1x siRNN conjugation buffer
- 5.25 M acetic acid
- 50 uM siRNNA4
- 1 mM DD-Hynic peptide (Note 3)
- 1x siRNN conjugation buffer
- Incubate reaction for 1 h at RT (Note 4).
- After 1 h, freeze the conjugation solution in crushed dry ice.
- Lyophilize the frozen conjugation solution until dry (Note 5).
- Once dry, dissolve the lyophilized pellet in 500 μl of ultrapure water by pipetting.
- Load 500 μl of crude DD-siRNN conjugate into a Amicon Ultra 0.5 ml centrifugal filter unit with Ultracel-30 membrane.
- Centrifuge the filled filter unit for 5 min at 14,000 x g.
- Discard flow-through.
- Add 450 μl of ultrapure water to the remaining liquid in the filter unit and pipette to mix.
- Repeat steps A7-9 two more times.
- Centrifuge the filled filter unit for 10 min at 14,000 x g.
- Invert the filter unit into a fresh collection tube and centrifuge for 2 min at 1,000 x g to elute purified DD-siRNN conjugate.
- Measure the absorbance of the purified DD-siRNN conjugate at 260 nm on a UV spectrophotometer.
- Determine the concentration of the purified DD-siRNN conjugate using the measured absorbance at 260 nm and the duplexed siRNN molar extinction coefficient (Notes 6 and 7).
- Dilute the purified DD-siRNN conjugate to the desired concentration with ultrapure water. Store DD-siRNN conjugate at -20 °C (Note 8).
- Set up the following conjugation reaction in a 1.5 ml microcentrifuge tube in 50% ACN /50% ultrapure water (Note 2):
- Analysis of DD-siRNN conjugation
- Purchase or cast 1.5 mm 0.4% SDS 10% polyacrylamide non-denaturing gels with 10 or 15-well combs (Note 9).
- Aliquot 0.1 nmol of purified DD-siRNN conjugates to be analyzed in fresh microcentrifuge tubes (Note 10).
- Freeze samples on crushed dry ice and lyophilize until dry (Notes 11 and 12).
- Once dry, dissolve lyophilized samples in 5 μl of 1x 0.4% SDS non-denaturing loading buffer by pipetting (Note 13).
- Place polymerized gels into the gel running apparatus and fill with 0.4% SDS-PAGE running buffer. Gently remove the combs from the gels and clean out the wells by forcefully pipetting running buffer into the empty wells.
- Load samples dissolved in loading buffer into wells of the gel and run at 200 V constant for 30 min or until the dye front runs off the gel.
- Once the gel has finished running, carefully separate the glass cassette and remove the gel. Deposit the gel in a small dish for staining.
- Rinse the gel several times with deionized water to remove running buffer (Note 14).
- Add 50-100 ml of deionized water to the staining dish and incubate the gel at room temperature on a rocker for 20 min (Note 15).
- Discard the wash and rinse the gel with deionized water.
- Conduct silver stain as described in the SilverQuest Silver Staining Kit protocol under “Basic Staining Protocol” (Notes 16 and 17).
- Image silver-stained gels on a Gel Doc to determine degree of conjugation and purity of samples (Figure 2B).
- Purchase or cast 1.5 mm 0.4% SDS 10% polyacrylamide non-denaturing gels with 10 or 15-well combs (Note 9).
- DD-siRNN cellular transduction and analysis
- Culture cells in desired media. For this protocol, H1299 cells stably expressing a destabilized EGFP (H1299-dGFP) were cultured in high glucose DMEM media, supplemented with 5% FBS and penicillin-streptomycin at 37 °C, 5% CO2.
- Rinse H1299 cells in PBS, trypsinize H1299 cells with 0.05% Trypsin-EDTA for 5 min, add growth media, and gently pipet up and down to dissociate cells from the culture dish.
- Centrifuge cells in bench centrifuge for 5 min at 200 x g.
- Discard supernatant, rinse cells in Opti-MEM, and centrifuge cells for 5 min at 200 x g.
- Resuspend cells in fresh Opti-MEM and dilute cells to 5 x 105 cells/ml with Opti-MEM.
- For each cell treatment, dilute DD-siRNN conjugates in 150 μl of fresh Opti-MEM (recommended final concentration of 50-500 nM). Transfer 150 μl of DD-siRNN dilution into a fresh well on a 24-well cell culture plate.
- Add 150 μl of H1299 cells (5 x 105 cells/ml) to wells containing diluted DD-siRNNs for a final concentration of 75,000 cells/well. Mix by carefully pipetting up and down several times.
- Incubate transductions at 37 °C, 5% CO2 for 2 h.
- At the end of the transduction time, aspirate off the treatment media and replace it with culture media (Note 18).
- After 24 h, trypsinize the transduced cells and split into three equal portions. Use two portions to seed fresh 24-well plates for later analysis of 48 and 72 h time-points.
- Analyze the final portion of cells on a flow cytometer to measure siRNN-induced knockdown of GFP (Figure 3A-B) (Note 19).
- Culture cells in desired media. For this protocol, H1299 cells stably expressing a destabilized EGFP (H1299-dGFP) were cultured in high glucose DMEM media, supplemented with 5% FBS and penicillin-streptomycin at 37 °C, 5% CO2.
Representative data
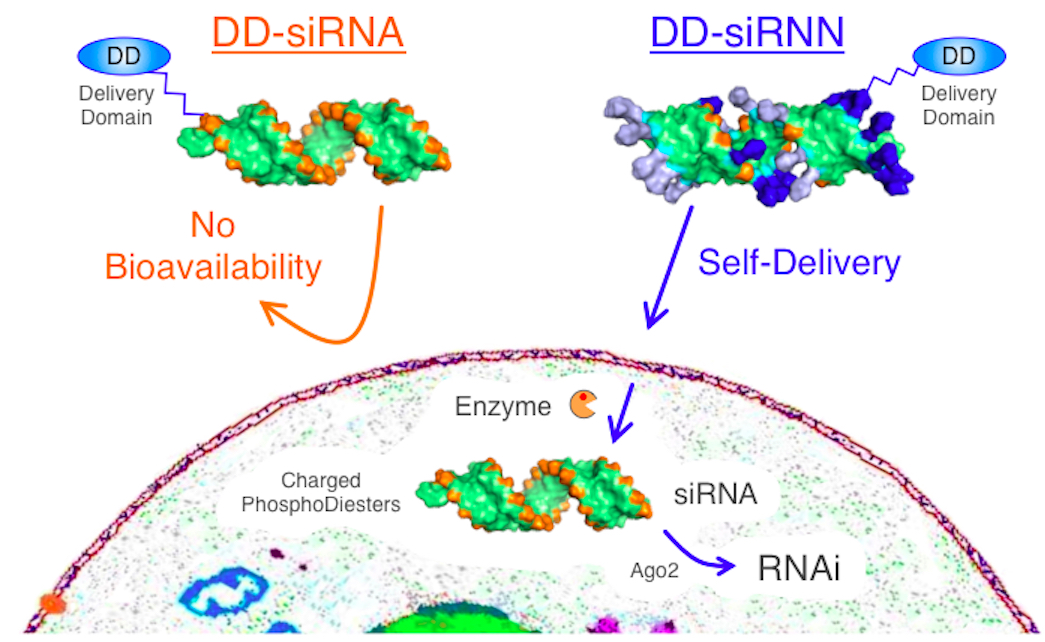
Figure 1. siRiboNucleic Neutrals (siRNNs) for cellular delivery of RNAi. siRNAs with charged phosphodiester backbones cannot cross the cell membrane unassisted. siRNNs feature bioreversible phosphotriesters which neutralize the phosphodiester backbone negative charge to allow for cellular delivery by TAT PTD delivery domain peptides and induction of RNAi responses. Source: S.F. Dowdy Lab, UCSD.
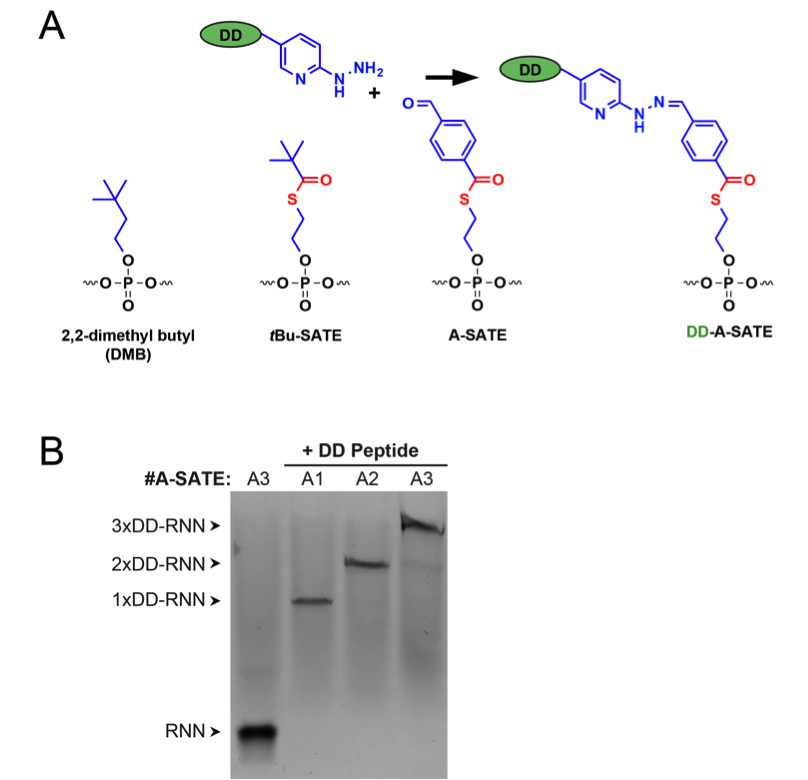
Figure 2. siRNN structure and conjugation with delivery domain (DD) peptides. A. Structures of phosphotriester groups used in this protocol. DD peptides containing hydrazine groups are conjugated to siRNNs through chemically reactive A-SATE phosphotriester group. Both the phosphotriester group and the conjugated peptide are removed by cleavage of DD-A-SATE by cytoplasmic thioesterases. B. Conjugation of DD peptides to RNN oligonucleotides containing one, two, or three A-SATE phosphotriester groups. Samples were resolved by SDS-PAGE and sliver-stained as described in this protocol. Source: S.F. Dowdy Lab, UCSD.
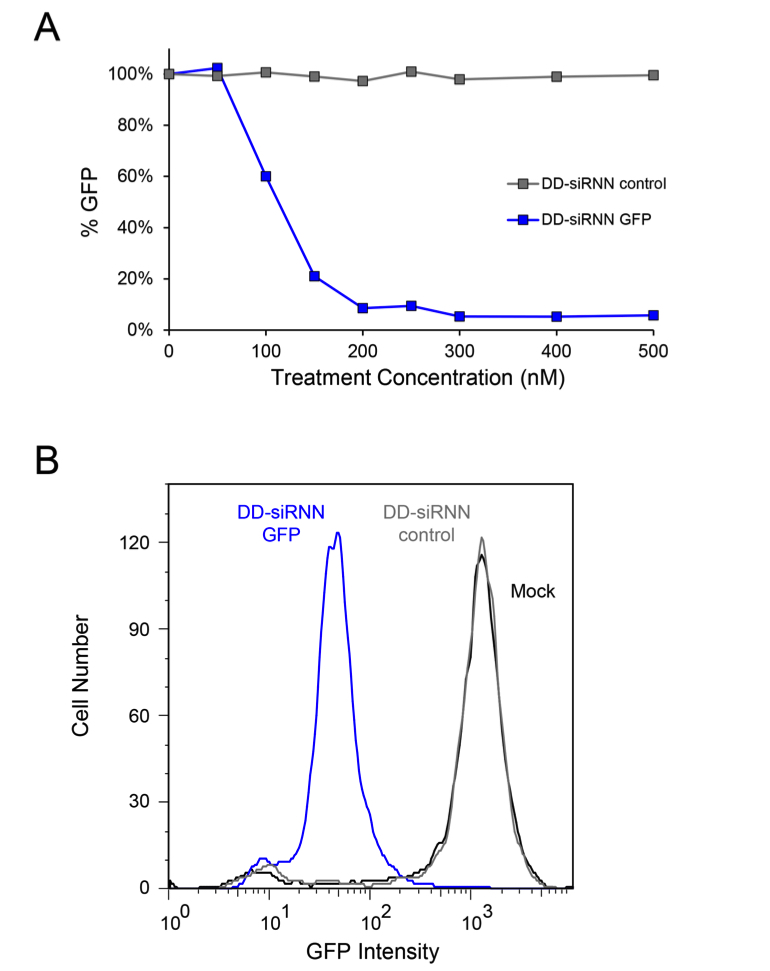
Figure 3. DD-siRNN cellular transduction. A. Dose curves of H1299-dGFP cells 48 h after treatment with self-delivering GFP DD-siRNNA4 or non-targeting control DD-siRNNA4. B. Histogram of H1299-dGFP cells analyzed by flow cytometry 48 h after treatment with GFP or control DD-siRNNA4 conjugates. As depicted here, the entire population undergoes an RNAi response following a successful transduction with on-target DD-siRNN conjugate. Source: S.F. Dowdy Lab, UCSD.
Notes
- The DD peptide (3T3S-Hy) used in this protocol was synthesized on site, but can be ordered from a variety of vendors with custom peptide synthesis services.
- This recipe is optimized for positively-charged DD peptide conjugation to siRNNs and may need to be adjusted to conjugate other molecules.
- Because there are 4 aldehyde A-SATE phosphotriester groups per molecule of siRNNA4 this results in a 5:1 ratio of DD peptide to A-SATE.
- For reaction volumes greater than 250 μl it is advantageous to conduct this reaction on a rotisserie.
- It is important that the conjugation solution be lyophilized until fully dry as residual acetic acid and ACN can distort the centrifugal filter membrane, preventing purification of DD-siRNN conjugates. The amount of time required to lyophilize a sample is dependent on the equipment used. A lyophilization rate of 50 μl/h can be expected using the equipment listed in this protocol with a vacuum pressure of 200 mTorr and a condenser temperature of -80 °C.
- Use the Beer-Lambert Law to calculate the concentration of the purified DD-siRNN conjugate. The Beer-Lambert Law: A=ϵlc where A is the measure of absorbance (at 260 nm in this case), ϵ is the molar extinction coefficient, l is the path length (1 cm for most spectrophotometers), and c is the concentration of the solution.
- The molar extinction coefficient for a duplexed oligonucleotide should be empirically determined. 292,500 (M-1 cm-1) is the molar extinction coefficient for the GFP siRNN used in this protocol.
- 20 μM is a useful concentration to dilute DD-siRNN conjugates for 24-well plate treatments.
- If casting your own gels make 0.4% SDS non-denaturing polyacrylamide gel solution in a 50 ml conical tube. Add 200 μl of 10% ammonium persulfate and 800 μl of 10% SDS solution to the gel solution. Mix by inverting the tube several times. Add 20 μl of TEMED to the gel solution and invert the tube several times to mix. Cast two 1.5 mm gels with the gel solution and glass plates. Carefully insert gel combs without introducing or trapping any air bubbles. Let gels sit at room temperature for 20 min to ensure complete polymerization before use.
- If DD-siRNN conjugates were diluted to 20 μM, 5 μl (0.1 nmol) should be aliquoted for analysis.
- Dry ice is extremely cold (-78.5 °C). Always handle dry ice with care and wear appropriate personal protective equipment (eye protection and insulated gloves).
- Lyophilization time will be dependent on the equipment and volume, but 30 min should be sufficient to fully dry 5 μl of DD-siRNN conjugate dissolved in water.
- Do not heat DD-siRNN conjugates as this may result in melting of the siRNN duplex. Once conjugated to a DD peptide, single-stranded RNNs (ssRNNs) will not readily duplex.
- A 12 x 10 x 3 cm polypropylene dish was used for staining in this protocol.
- This step decreases background staining during the silver stain by washing some SDS out of the gel.
- The SilverQuest Silver Staining Kit manual recommends using 100 ml for all solutions, but 50 ml may be used instead as long as the staining dish is small enough for 50 ml to completely cover the gel.
- If 0.1 nmol of DD-siRNN conjugate is loaded, 1-5 min of silver stain development time should be sufficient to visualize the conjugate.
- Under these conditions, H1299 cells will adhere to the plate after 2 h. If the cells do not adhere sufficiently for complete aspiration of the transduction media, add 500 μl of culture media, incubate the plate until the cells adhere, and then aspirate all of the media.
- For this protocol, H1299-dGFP cells were analyzed on a LSR II flow cytometer. dGFP was measured using a 488 nm laser and a 530/30 emission filter. 5 x 104 events were collected and gated according to forward scatter (FSC-A) and side scatter (SSC-A).
- Abbreviations used in this protocol: siRNA = short interfering RNA, PTD = peptide transduction domain, siRNN = siRiboNucleic Neutral, DMB = dimethyl butyl, SATE = S-acyl-2-thioethyl, HyNic = 6-hydrazinonicotinamide.
Recipes
- 5x siRNN conjugation buffer (1 ml)
500 μl acetonitrile
315.5 μl ultrapure water
9.5 μl 10.5 M aniline (100 mM final concentration)
125 μl of 200 mM HEPES (pH 5.5) (25 mM final concentration)
50 μl 5 M NaCl (250 mM final concentration) - 0.4% SDS non-denaturing polyacrylamide gel solution
12.2 ml of ultrapure water
5 ml 40% acrylamide/bis solution
2 ml 10x TBE - 0.4% SDS-PAGE running buffer (1 L)
40 ml of 10% SDS solution
100 ml of 10x TBE
860 ml deionized water - 1x 0.4% SDS non-denaturing loading buffer (1 ml)
40 μl 10% SDS solution
250 μl glycerol
2 mg Orange G
Add ultrapure water to 1 ml
Acknowledgments
The protocol described herein was developed and utilized previously in Meade et al. (2014). A. S. H. was supported by a T32 Cancer Biology Training grant (NCI) and by a Blasker Award from The San Diego Foundation. This work was supported by the W.M. Keck Foundation (S. F. D.), the Department of Defense (S. F. D.), SCOR grant from the Leukemia & Lymphoma Society (S. F. D.), the Pardee Foundation (S. F. D.), a grant from an anonymous donor (S. F. D.) and the Howard Hughes Medical Institute (S. F. D.).
The authors declare competing financial interests. S. F. D. and K.G. (UCSD) have filed patents on this work that were licensed by Solstice Biologics, Inc. (San Diego). S. F. D. is a cofounder of Solstice Biologics. S. F. D. is a board director of Solstice Biologics.
References
- Bumcrot, D., Manoharan, M., Koteliansky, V. and Sah, D. W. (2006). RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol 2(12): 711-719.
- Farkhani, S. M., Valizadeh, A., Karami, H., Mohammadi, S., Sohrabi, N. and Badrzadeh, F. (2014). Cell penetrating peptides: efficient vectors for delivery of nanoparticles, nanocarriers, therapeutic and diagnostic molecules. Peptides 57: 78-94.
- Meade, B. R. and Dowdy, S. F. (2007). Exogenous siRNA delivery using peptide transduction domains/cell penetrating peptides. Adv Drug Deliv Rev 59(2-3): 134-140.
- Meade, B. R., Gogoi, K., Hamil, A. S., Palm-Apergi, C., van den Berg, A., Hagopian, J. C., Springer, A. D., Eguchi, A., Kacsinta, A. D., Dowdy, C. F., Presente, A., Lonn, P., Kaulich, M., Yoshioka, N., Gros, E., Cui, X. S. and Dowdy, S. F. (2014). Efficient delivery of RNAi prodrugs containing reversible charge-neutralizing phosphotriester backbone modifications. Nat Biotechnol 32(12): 1256-1261.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Hamil, A. S., Gogoi, K. and Dowdy, S. F. (2016). Conjugation of Duplexed siRNN Oligonucleotides with DD-HyNic Peptides for Cellular Delivery of RNAi Triggers. Bio-protocol 6(7): e1782. DOI: 10.21769/BioProtoc.1782.
Category
Molecular Biology > RNA > RNA interference
Molecular Biology > RNA > RNA synthesis
Biochemistry > RNA > RNA structure
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









