- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Total RNA Extraction from Grape Berry Skin for Quantitative Reverse Transcription PCR and Microarray Analysis
Published: Vol 6, Iss 7, Apr 5, 2016 DOI: 10.21769/BioProtoc.1777 Views: 8914
Reviewed by: Tie LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

RNA Purification from the Unicellular Green Alga, Chromochloris zofingiensis
Sean D. Gallaher and Melissa S. Roth
Apr 5, 2018 8157 Views

Extraction of RNA from Recalcitrant Tree Species Paulownia elongata
Niveditha Ramadoss and Chhandak Basu
Jul 20, 2018 7723 Views

Simple Method for Efficient RNA Extraction From Arabidopsis Embryos
Fernanda Marchetti [...] Eduardo Zabaleta
Feb 20, 2025 1927 Views
Abstract
Extraction of high quality RNA is an essential step for quantitative reverse transcription PCR (qRT-PCR) and microarray analysis. However, it is not easy to extract high quality RNA from fruit materials, which contain high amounts of polysaccharides, lipids and secondary metabolites. Wan and Wilkins (1994) had developed ‘Hot Borate Method’ to isolate high quality RNA. Here, we describe a modified protocol of the ‘Hot Borate Method’ to isolate high quality RNA from grape berry skin for qRT-PCR and microarray analysis (Suzuki et al., 2015a; Suzuki et al., 2015b).
Keywords: RNA extractionMaterials and Reagents
- 50 ml centrifuge tubes (e.g., the Falcon tubes)
- 1.5 ml microcentrifuge tubes
- 2 ml microcentrifuge tubes
- Stainless steel spoon
- Grape berry skins
Notes:- Skins isolated from grape berries using tweezers (Figure 1A, Video 1) . Skins were put in 15 or 50 ml tubes and frozen in liquid nitrogen immediately (Figure 1B, Video 1). These were stored at -80 °C. We recommend that preparation of about 20 pieces (about 1 g skin) because there are variations among berries.
- This method could be applied to all grape varieties.
- Skins isolated from grape berries using tweezers (Figure 1A, Video 1) . Skins were put in 15 or 50 ml tubes and frozen in liquid nitrogen immediately (Figure 1B, Video 1). These were stored at -80 °C. We recommend that preparation of about 20 pieces (about 1 g skin) because there are variations among berries.
- Liquid nitrogen
- Proteinase K solutions (20 mg/ml) (Siyaku, Wako Pure Chemical Industries, catalog number: 160-22752 )
- Lithium chloride (LiCl) (2 M, 10 M) (Siyaku, Wako Pure Chemical Industries, catalog number: 127-01165 )
Note: DEPC treated and autoclaved. - 2 M potassium acetate (KOAC) (Siyaku, Wako Pure Chemical Industries, catalog number: 160-03175 )
Note: DEPC treated and autoclaved. - 10 mM Tris-HCl (pH 7.5) (Siyaku, Wako Pure Chemical Industries, catalog number: 207-06275 )
Note: DEPC treated and autoclaved. - 1 M potassium chloride (KCl) (Siyaku, Wako Pure Chemical Industries, catalog number: 163-03545 )
Note: DEPC treated and autoclaved. - Ethanol [70%, 99.8% (v/v)] (Siyaku, Wako Pure Chemical Industries, catalog number: 057-00456 )
- RNeasy Plant Mini Kit (QIAGEN, catalog number: 74903 or 74904 )
- RNase free water
- Sodium borate decahydrate (Borax) (Siyaku, Wako Pure Chemical Industries, catalog number: 194-01415 )
- Ethylene glycol tetraacetic acid (EGTA) (Dojindo, catalog number: 346-01312 )
- 1% (w/v) sodium dodecyl sulfate (SDS) (Siyaku, Wako Pure Chemical Industries, catalog number: 191-07145 )
- 1% (w/v) deoxycholate sodium salt (Siyaku, Wako Pure Chemical Industries, catalog number: 192-08312 )
- 10 mM dithiothreitol (DTT) (Siyaku, Wako Pure Chemical Industries, catalog number: 045-08974 )
- 1% (w/v) Triton X-114 (Sigma-Aldrich, catalog number: X114 )
- 2% (w/v) polyvinylpyrrolidone (PVP-40) (Sigma-Aldrich, catalog number: PVP40 )
- Diethyl pyrocarbonate (DEPC) (Sigma-Aldrich, catalog number: D5758 ) treated and autoclaved
- Hot Borate Extraction Buffer (see Recipes)
Equipment
- Crusher (Automill) (Tokken, model: TK-AM5-H )
- Shaking water bath (42 °C) (TAITEC CORPORATION, model: THERMO MINDER SM-05 with PERSONAL-11)
- Centrifuge with angle rotor for 50 ml tube (SAKURA, model: 50A-7 with 50F-8A)
- Centrifuge with angle rotor for 1.5 ml tube (SAKURA, model: SS1500X with 15M-24AM)
- Vacuum equipment (EYELA, model: CVE-2000 )
Procedure
- Extraction of RNA from the sample
- Grape berry skins are prepared as shown in Figure 1 and Video 1.
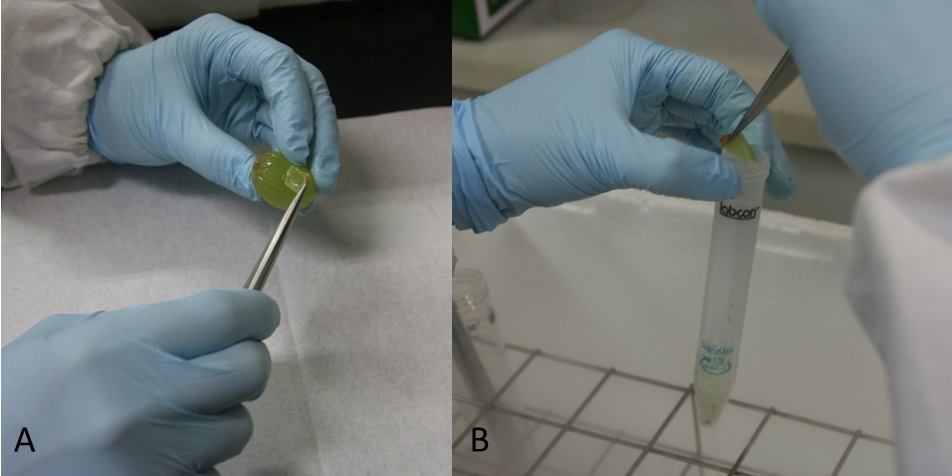
Figure 1. Preparation of grape berry skins. Grape skins were separated from berry (A) and frozen in liquid nitrogen (B)Video 1. Preparation of grape berry skins - Powder frozen grape berry skins with liquid N2 using the crusher (1,300 rpm, 30 sec) (Figure 2A-B, Video 2).
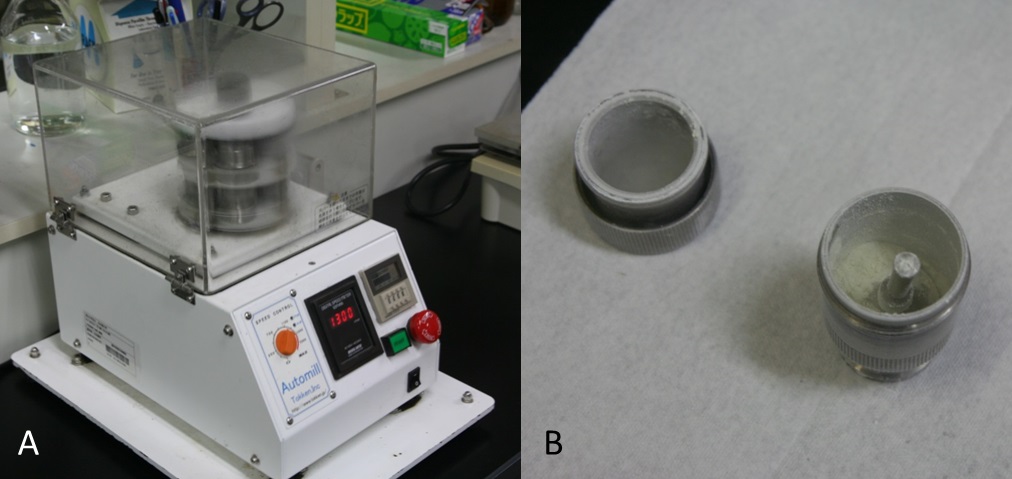
Figure 2. The crusher (A) and powdered grape berry skins (B)Video 2. Powder frozen grape berry skins with liquid N2 using the crusher - Put 0.2 g powdered tissue into 50 ml centrifuge tube using the cold spoon (Figure 3A, Video 3).
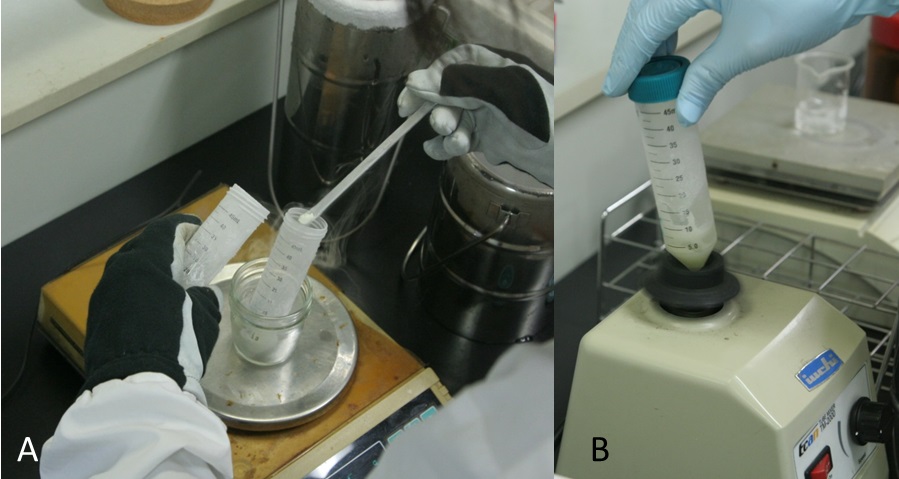
Figure 3. Preparation of RNA sample. A. Powdered tissue was put into tube using the cold spoon; B. The sample was mixed by vortexing.Video 3. Preparation of RNA sample - Add 2 ml hot borate extraction buffer (42 °C) (Video 3).
- Add 10 μl Proteinase K solutions (20 mg/ml) immediately (Video 3).
- Mix the sample by vortexing until well mixed (approximately 20 sec) (Figure 3B, Video 3).
- Incubate the tube at 42 °C in the shaking water bath for 1.5 h.
- Centrifuge at 8,000 rpm (8,000 x g) for 10 min at room temperature.
- Transfer the supernatant to three 1.5 ml tubes (600 μl/tube).
- Add 114.3 μl of 1 M KCl to each tube to adjust a final concentration of 160 mM.
- Incubate the tubes on ice for 1 h.
- Centrifuge at 10,000 rpm (12,000 x g) for 30 min at 4 °C to remove debris.
- Transfer the supernatants to 1.5 ml microcentrifuge tube individually.
- Discard pellet and use transferred supernatant in step B (see below).
- Grape berry skins are prepared as shown in Figure 1 and Video 1.
- Purification of RNA (remove polysaccharide)
- Add 178.5 μl of 10 M LiCl to each tube to adjust a final concentration of 2 M.
- Incubate on ice in a cold room overnight.
- Centrifuge at 10,000 rpm (12,000 x g) for 30 min at 4 °C.
- Decant and discard the supernatant immediately.
- Add 200 μl of cold 2 M LiCl for each tube.
- Suspend the pellet by vortexing until well mixed (approximately 20 sec).
- Centrifuge at 10,000 rpm (12,000 x g) for 10 min at 4 °C.
- Decant and discard the supernatant immediately.
- Repeat steps B5-8 twice (for a total of three times).
- If the pellet contains pigment, repeat steps B5-8 until the pellet becomes colorless.
- Suspend the pellet in 100 μl of 10 mM Tris-HCl (pH 7.5) at room temperature.
- Collect the suspensions in one 1.5 ml microcentrifuge tube and adjust to 400 μl with 10 mM Tris-HCl (pH 7.5).
- Centrifuge at 10,000 rpm (12,000 x g) for 10 min at 4 °C to remove insoluble materials.
- Transfer the supernatant to 1.5 ml microcentrifuge tube.
- Add 40 μl of 2 M KOAC to adjust a final concentration of 0.2 M.
- Incubate on ice for 15 min.
- Centrifuge at 10,000 rpm (12,000 x g) for 10 min at 4 °C.
- Transfer the supernatant to 2 ml microcentrifuge tube.
- Add 1.1 ml of 99.8% ethanol and keep at -20 °C overnight to precipitate RNA.
- Add 178.5 μl of 10 M LiCl to each tube to adjust a final concentration of 2 M.
- Cleanup and concentration of RNA
- Centrifuge at 10,000 rpm (12,000 x g) for 30 min at 4 °C to pellet RNA.
- Decant and discard ethanol.
- Wash pellet with 70% (v/v) ethanol.
- Centrifuge at 10,000 rpm (12,000 x g) for 10 min at 4 °C.
- Decant and discard ethanol.
- Remove residual ethanol under vacuum.
- Suspend the pellet in 100 μl RNase free water.
- Stored at -80 °C until use for qRT-PCR or keep at 4 °C until further purification.
- Centrifuge at 10,000 rpm (12,000 x g) for 30 min at 4 °C to pellet RNA.
- Further cleanup and concentration of RNA for microarray analysis
- The RNA was further purified using RNeasy Plant Mini Kit according to the supplier’s protocol.
- Add 350 μl of RLT buffer and mix thoroughly.
- Add 250 μl of 99.8% ethanol and mix well by pipetting.
- Apply sample to RNeasy mini spin column sitting with collection tube.
- Centrifuge at 8,000 rpm (8,000 x g) for 15 sec.
- Transfer the column to a new tube.
- Add 500 μl of RPE buffer and centrifuge at 8,000 rpm (8,000 x g) for 15 sec.
- Add 500 μl of RPE buffer and centrifuge at 8,000 rpm (8,000 x g) for 2 min.
- Discard flow-through and transfer the column to a new tube.
- Centrifuge at 10,000 rpm (12,000 x g) for 1 min.
- Discard flow-through and transfer the column to a new tube.
- Add 100 μl of water and centrifuge at 8,000 rpm (8,000 x g) for 1 min.
- Add 100 μl of water and centrifuge at 8,000 rpm (8,000 x g) for 1 min.
- Collect the filtrates and add 500 μl of 99.8% ethanol.
- Keep at -20 °C overnight to precipitate RNA.
- Repeat steps C1-6.
- Suspend the pellet in 10 μl RNase free water.
- Stored at -80 °C until use for microarray analysis.
- The RNA was further purified using RNeasy Plant Mini Kit according to the supplier’s protocol.
Recipes
- Hot borate extraction buffer
0.2 M sodium borate decahydrate (Borax)
30 mM ethylene glycol tetraacetic acid (EGTA)
1% (w/v) sodium dodecyl sulfate (SDS)
1% (w/v) deoxycholate sodium salt
10 mM dithiothreitol (DTT)
1% (w/v) Triton X-114
2% (w/v) polyvinylpyrrilidone (PVP-40)
Notes:- Store Borax, EGTA, SDS and deoxycholate sodium salt as 1.5x mixture after autoclave.
- DTT was filtered and stored as 1 M solution at -80 °C.
- Store PVP-40 as 20% (w/v) solution after autoclave.
- Add DTT, Triton X-114 and PVP-40 before use.
- Store Borax, EGTA, SDS and deoxycholate sodium salt as 1.5x mixture after autoclave.
Acknowledgments
This work was supported by the Programme for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry from the Bio-oriented Technology Research Advancement Institution (BRAIN) and by Grants-in-Aid for Scientific Research from The Japan Society for the Promotion of Science (JSPS).
We thank Dr. Wan and Dr. Wilkins for showing method of ‘Hot Borate Method’.
We also thank Dr. Azuma and Mr. Nakao for helping us making of figures and videos.
References
- Suzuki, M., Jasinski, M., Martinoia, E., Nakabayashi, R., Suzuki, M., Saito, K. and Shiratake, K. (2015a). Molecular cloning and characterization of ABCG/PDRtype ABC transporter in grape berry skin. Adv Hortic Sci 28: 53-63.
- Suzuki, M., Nakabayashi, R., Ogata, Y., Sakurai, N., Tokimatsu, T., Goto, S., Suzuki, M., Jasinski, M., Martinoia, E., Otagaki, S., Matsumoto, S., Saito, K. and Shiratake, K. (2015b). Multiomics in grape berry skin revealed specific induction of the stilbene synthetic pathway by ultraviolet-C irradiation. Plant Physiol 168(1): 47-59.
- Wan, C. Y. and Wilkins, T. A. (1994). A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.). Anal Biochem 223(1): 7-12.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Suzuki, M. and Shiratake, K. (2016). Total RNA Extraction from Grape Berry Skin for Quantitative Reverse Transcription PCR and Microarray Analysis. Bio-protocol 6(7): e1777. DOI: 10.21769/BioProtoc.1777.
- Suzuki, M., Nakabayashi, R., Ogata, Y., Sakurai, N., Tokimatsu, T., Goto, S., Suzuki, M., Jasinski, M., Martinoia, E., Otagaki, S., Matsumoto, S., Saito, K. and Shiratake, K. (2015b). Multiomics in grape berry skin revealed specific induction of the stilbene synthetic pathway by ultraviolet-C irradiation. Plant Physiol 168(1): 47-59.
Category
Plant Science > Plant molecular biology > RNA > RNA extraction
Molecular Biology > RNA > RNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












