- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Immunogold Labeling Analysis of Cell Wall Polysaccharides with Special Reference to (1;3,1;4)-β-D-glucan in Rice Cell Walls
Published: Vol 6, Iss 5, Mar 5, 2016 DOI: 10.21769/BioProtoc.1748 Views: 9433
Reviewed by: Tie LiuCarsten AdeValérie Cornuault

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Enzymatic Starch Quantification in Developing Flower Primordia of Sweet Cherry
Nestor Santolaria [...] Afif Hedhly
Apr 5, 2025 1897 Views

New Approach to Detect and Isolate Rhamnogalacturonan-II in Arabidopsis thaliana Seed Mucilage
Dayan Sanhueza and Susana Saez-Aguayo
Sep 5, 2025 1248 Views

Detailed Method for the Purification of Rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana
Liang Zhang [...] Breeanna R. Urbanowicz
Feb 5, 2026 144 Views
Abstract
Various types of cell wall compositions have evolved to fulfill a wide range of biological roles during the diversification of land plants. (1;3,1;4)-β-D-glucan (MLG) is a defining feature of the cell walls in the order Poales (Yokoyama and Nishitani, 2004), which has multiple functions associated with metabolic, growth, and defense systems. MLG is also a characteristic component of the matrix polysaccharides that undergo turnover and metabolism, depending on the tissue and the stage of development (Kido et al., 2015). Determining the extracellular localization of MLG is essential for elucidating its functions. Electron microscopy immunogold labeling analysis is a useful technique, which provides an accurate representation of the extracellular distribution of MLG. This strategy is also applicable to various kinds of cell wall polysaccharides, which have key roles in regulating growth and differentiation in each plant species.
Keywords: Electron microscopyMaterials and Reagents
- Gelatin capsule (HF capsule) (Matsuya Co.)
- Rice plants (see Note)
- Paraformaldehyde (Electron Microscopy Sciences, catalog number: 12300 )
- Sodium cacodylate buffer solution (TAAB Laboratories BATCH, catalog number: 90760 )
- Glutaraldehyde (Electron Microscopy Sciences, catalog number: 16220 )
- Tannic acid (Sigma-Aldrich, catalog number: 403040 )
- Phosphate buffer powder (Siyaku, Wako Pure Chemical Industries, catalog number: 167-14491 )
- Ethanol (Siyaku, Wako Pure Chemical Industries, catalog number: 057-00456 )
- LR White resin (Electron Microscopy Sciences, London Resin Company, catalog number: 14381-CA )
- Monoclonal antibody against (1;3,1;4)-β-D-glucan (mouse IgG, Kappa Light) (Biosupplies, catalog number: 400-3 )
Note: It is named as (1-3;1-4)-β-D-glucan on Biosupplies’ website. - Goat Anti-mouse IgG pAb, Gold 15 nm, EM (BBI Solutions, catalog number: EM.GMHL15 )
- Phosphate-buffered salts (Takara Bio Company, catalog number: T900 )
- Bovine serum albumin (BSA) (Sigma-Aldrich, catalog number: A2058 )
- Uranyl acetate (Siyaku, Wako Pure Chemical Industries, catalog number: 554-85072 )
- Lead stain solution (Sigma-Aldrich, catalog number: 18-0875-2-25 ml-J )
Note: This reagent may not be available outside Japan, and can be replaced with alternative lead stains. For an alternative, readers are referred to a more general experimental manual for electron microscopy of plant cells (Hall and Hawes, 1991). - Molecular sieves (Sigma-Aldrich, catalog number: M6141 )
- Fixative solution (see Recipes)
- 2% glutaraldehyde solution (see Recipes)
- 100% ethanol solution (see Recipes)
- 1x Phosphate-buffered saline (PBS) (see Recipes)
Equipment
- Growth chamber (Nippon Medical & Chemical Instruments, model: LH220S )
- Ultracut UCT Ultramicrotome (Leica Microsystems)
- Diaphragm vacuum pump (Leybold-Heraus, model: Divac 2.2 L )
- Desiccator (Sanplatec, model: PC-250K )
- Grid mesh (Okenshoji, catalog number: 09-1079 )
- Transmission electron microscope (JEOL USA, model: JEM-1400plus )
- CCD camera (Olympus soft imaging solutions GmbH, model: Veleta )
Procedure
- Cut the rice leaf blade into pieces measuring ≤ 2 mm in length and immediately immerse them in a fixative solution. The fixative solution should be handled under a fume hood.
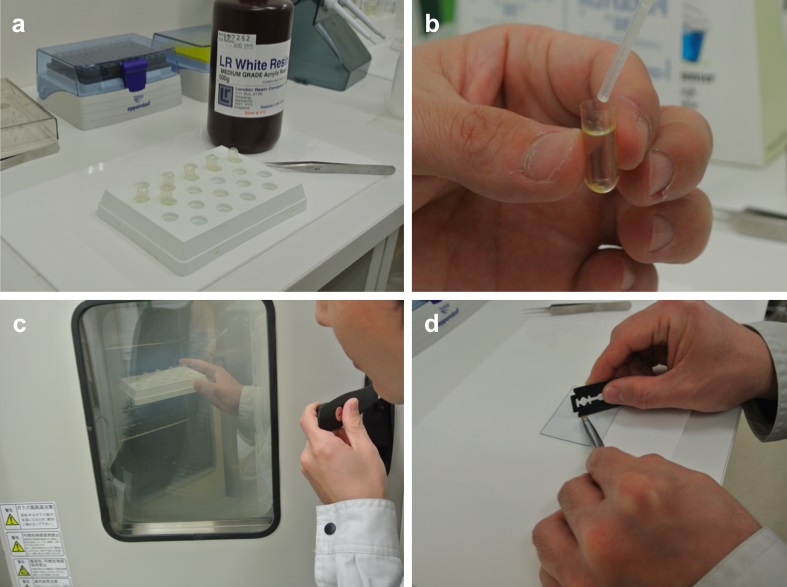
Figure 1. Embedding and trimming. a. Equipment and reagent. b. The tissues in the bottom of gelatin capsules filled with fresh 100% LR White resin. c. Polymerization. d. Trimming. - The tissue/fixative is placed in a desiccator, which is connected to a diaphragm vacuum pump. The tissue/fixative is vacuum degasified for 1 min at room temperature using the vacuum pump before releasing the vacuum very slowly.
- Incubate the tissue/fixative for 2 h at 4 °C.
- Rinse the tissues gently three times with 50 mM sodium cacodylate buffer (pH 7.4) for 15 min each time.
- The tissues are dehydrated using a graded ethanol series (70%, 80%, 90%, and three times at 100%) at 4 °C for 30 min in each solution. Swirl occasionally.
- Immerse the tissues three times with a 50:50 mixture of 100% (v/v) ethanol and 100% (v/v) LR White resin for 30 min each time at room temperature.
- Transfer the tissues to 100% LR White resin and immerse the tissues three times for 1 h each time at room temperature. This step ensures that no residual ethanol will be carried over since other chemical like ethanol or else prevents polymerization of LR resin.
- The tissues are settled to the bottom of gelatin capsules filled with fresh 100% LR White resin (Figure 1b). Stand the gelatin capsules in the test-tube rack for polymerization overnight at 50 °C (Figure 1c).
- Turn the tissue block upside down and secure it in a UCT ultramicrotome. Chuck and trim the block to expose the tissue surface area up to 1.0 sq mm using a razor blade (Figure 1d).
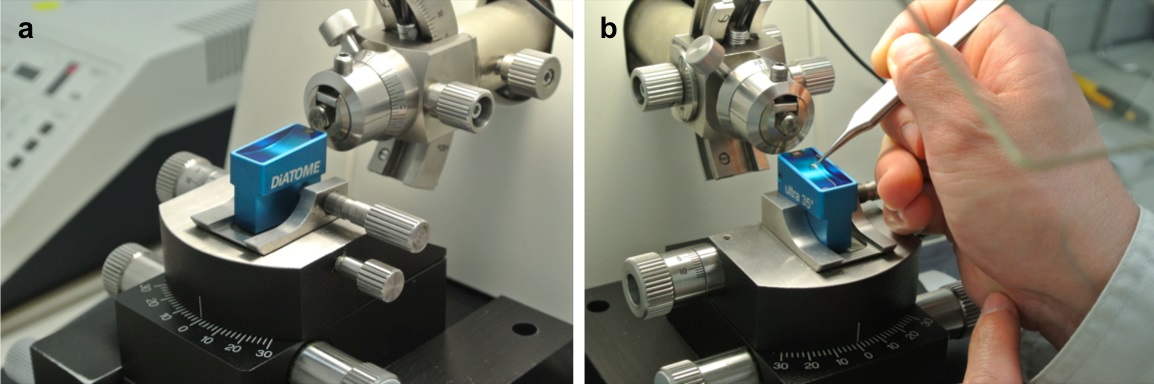
Figure 2. Sectioning. a. Sectioning with a ultramicrotome. b. Mounting the ultrathin sections on the grid. - Cut the block to 80 nm with a diamond knife on a UCT ultramicrotome (Figure 2a). Dip up the serial sections floating on water in the boat of the diamond knife with a nickel grid mesh, and mount the ultrathin sections on the grid (Figure 2b).
- Incubate the grid with a droplet of the mouse anti-(1;3,1;4)-β-D-glucan antibody (diluted 1:1,000 in a 1% BSA solution, depending on the abundance of the target primary antibody) at room temperature for 90 min (Figure 3a).
- Rinse the grid three times with 1% BSA/1x PBS for 1 min each time.
- Float the grid on a droplet of goat anti-mouse IgG coupled to 15-nm gold particles 1:20 in 1% BSA/1x PBS at room temperature for 1 h. When detecting another primary antibody, prepare goat anti-mouse IgG coupled to gold particles to match the primary antibody.
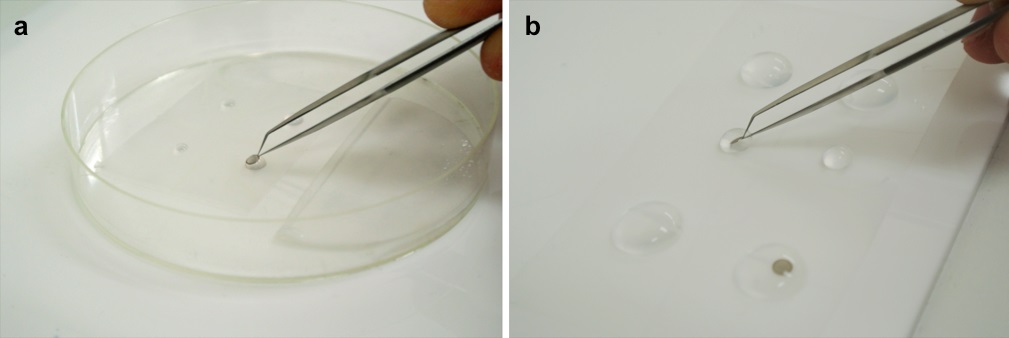
Figure 3. Immunostaining (a) and staining with uranyl acetate (b) - Rinse the grid and the sections three times with 1x PBS.
- Immerse the grid and the sections in 2% glutaraldehyde solution and are air-dried under a fume hood.
- Immerse the sections in a droplet of 2% uranyl acetate, which had been prepared by diluting with distilled water, for 15 min (Figure 3b).
- Rinse the sections with distilled water.
- Immerse the sections with Lead stain solution at room temperature for 3 min. Rinse the sections with distilled water.
- Observe the sections using a JEOL JEM 1200EX transmission electron microscope at an accelerating voltage of 80 kV.
- Digital images (2,048 x 2,048 pixels) are obtained with a CCD camera.
Representative data
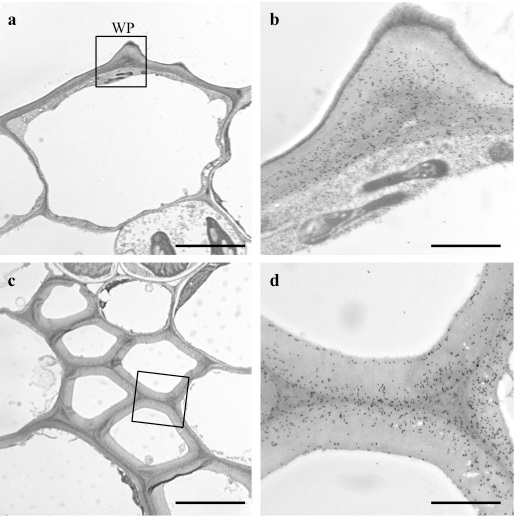
Figure 4. Immunogold-based localization of MLG is shown in the leaf blades. a. The images in (a) are of the cell walls of the epidermal cell with a wart-like protuberance (WP). Scale bar = 5 μm. b. A higher magnification of the rectangle in (a). Scale bar = 1 μm. c. The images in (c) are of the cell walls of the cortical sclerenchyma. Scale bar= 5 μm. d. A higher magnification of the rectangle in (c). Scale bar = 1 μm.
Notes
Rice (Oryza sativa L. cv Nipponbare) plants are grown in a growth chamber at 28 °C under a 15/9 h light/dark cycle (light at 150 μmol-2 s-1). This protocol is performed using the fifth leaf blades of the rice plants.
Recipes
- Fixative solution
Prepare fixative solution by dissolving 0.4 g of paraformaldehyde, 0.05 g of tannic acid, and 0.1 ml of 10% glutaraldehyde in 9.9 ml of 50 mM sodium cacodylate buffer (pH 7.4). Heat this solution in the fume hood on the hotplate/stirrer to approx. 70 °C until the solution clears. - 2% glutaraldehyde solution
Dissolve an appropriate volume of Wako Pure Chemical Industries ready-made powder in distilled water according to manufacturer's protocol, to make a 0.1 M phosphate buffer of pH 7.4 at room temperature. Prepare 2% glutaraldehyde solution by diluting 10% glutaraldehyde in the phosphate buffer. - 100% ethanol solution
To prepare the 100% ethanol solution, use 100% bulk ethanol with molecular sieves in the bottom of the bottle - 1x PBS
Dissolve 10 PBS tablets in distilled water to make a total volume of 1,000 ml
Acknowledgments
The electron microscopic analysis was performed by Tokai Electron Microscopy (Nagoya, Japan). This study was supported by the Japan Society for Promotion of Science (JSPS) and the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan [a Grant-in-Aid for Scientific Research on Innovative Areas “Plant Cell Wall as Information Processing System” (No. 24114001, 24114005) to K. N. and a Grant-in-Aid for Scientific Research (C) (25440124) to R.Y.].
References
- Hall, J. L. and Hawes, C. R. (1991). Electron microscopy of plant cells. Academic Press.
- Kido, N., Yokoyama, R., Yamamoto, T., Furukawa, J., Iwai, H., Satoh, S. and Nishitani, K. (2015). The matrix polysaccharide (1;3,1;4)-beta-D-glucan is involved in silicon-dependent strengthening of rice cell wall. Plant Cell Physiol 56(2): 268-276.
- Yokoyama, R. and Nishitani, K. (2004). Genomic basis for cell-wall diversity in plants. A comparative approach to gene families in rice and Arabidopsis. Plant Cell Physiol 45(9): 1111-1121.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Yokoyama, R., Kido, N., Yamamoto, T., Furukawa, J., Iwai, H., Satoh, S. and Nishitani, K. (2016). Immunogold Labeling Analysis of Cell Wall Polysaccharides with Special Reference to (1;3,1;4)-β-D-glucan in Rice Cell Walls. Bio-protocol 6(5): e1748. DOI: 10.21769/BioProtoc.1748.
Category
Plant Science > Plant biochemistry > Carbohydrate
Biochemistry > Carbohydrate > Polysaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









