- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurement of 33P-PO4 Absorption Capacity and Root-to-leaf Transfer in Arabidopsis
Published: Vol 6, Iss 5, Mar 5, 2016 DOI: 10.21769/BioProtoc.1741 Views: 7437
Reviewed by: Tie LiuMichael O. AduAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Visualization of Nitric Oxide, Measurement of Nitrosothiols Content, Activity of NOS and NR in Wheat Seedlings
Sandeep B. Adavi [...] Shailendra K. Jha
Oct 20, 2019 6354 Views

A Quick Method to Quantify Iron in Arabidopsis Seedlings
Chandan Kumar Gautam [...] Wolfgang Schmidt
Mar 5, 2022 3932 Views

CAPS-Based SNP Genotyping for Nitrogen-Response Phenotypes in Maize Hybrids
Jannis Jacobs [...] Peter K. Lundquist
Dec 20, 2025 550 Views
Abstract
This method allows quantification of phosphate absorption capacity by Arabidopsis roots using very simple equipment, and can be scaled up or down.
Keywords: Phosphate absorptionMaterials and Reagents
- Plastic 12-well plates (Denmark, Nunc)
Note: One for the absorption step, one for the desorption step for each treatment (plant type or culture condition). - Plastic 20 ml vials for radioactivity measurement (Ratiolab GmbH, Dreieich)
Note: You will need one vial per plant, or 2 vials per plant if you want to quantify 33P in both roots and leaves. The vials should be numbered from 1 to N before you start the experiment. They also should be placed in order in appropriate racks (PerkinElmer) adapted to the beta counter. - Tips
- Young in vitro plantlets
- MES hydrate (Sigma-Aldrich, catalog number: M8250 )
- CaCl2 (Sigma-Aldrich)
- KH2PO4 (Sigma-Aldrich)
- 33P-PO4 5 mCi/ml (40-158 Ci/mg, 1.48-5.84 TBq/mg, >99% isotopically pure, less than 0.5 μM Pi) (PerkinElmer)
- Scintillation cocktail (PerkinElmer, Ultima GoldTM)
- MgSO4
- NH4NO3
- KNO3
- NaH2PO4
- KI
- FeCl2
- MnSO4
- ZnSO4
- CuSO4
- CoCl2
- Na2MoO4
- Thiamine
- Pyridoxine
- Nicotinic acid
- Inositol
- Sucrose
- Agar
- MS/10 medium (see Recipes)
- Stock solution (see Recipes)
- Incubation medium (see Recipes)
- Desorption medium (see Recipes)
Equipment
- Experiments should be performed on a bench or under a hood illuminated with white light (150 -180 μE m-2 s-1) during the incubation step
- Liquid scintillation counter (PerkinElmer, Packard Instrument Company, model: TRI-CARB )
- Ice-containing large boxes for the desorption step (all 12-well plates will be placed horizontally on ice for 2 h)
- Tweezers for handling the plantlets
- If necessary, a razor to separate roots and aerial parts
- Micropipets
- Shield for protection against radiations (plexiglass)
- Scanner or camera (Epson America, model: Perfection V850Pro or Canon, model: Powershot SX130 ), respectively but other devices from other manufacturers could suit perfectly
Software
- ImageJ version 1.46r with NeuronJ plugin (http://imagej.nih.gov/ij)
Procedure
- Preparation of plant materials
9 to 11 day-old plantlets are convenient for the experiment.- When plating, space the seeds in order to maintain root systems independent for every single plantlet; this will help when measuring the length of the root system (see below). 6 to 10 seeds are sown per plate depending on the growth medium (respectively with high or low Pi, see below). 2 to 3 plates will be necessary per plant type (genotype or treatment depending on the experiment).
- Plants are grown vertically in 12 x 12 cm Petri plates in modified MS/10 medium.
- Number the plants on each Petri plate, in order to identify each root system individually.
- 10 to 12 replicates per treatment or genotype will be necessary plus 5 to 10 plants for blanks.
- Scan or photograph the plates (in black and white and jpg format). Use graph paper as scale bar for measurement of the root length. Scan should be 300 dpi or photos should be about 1,000 x 800 pixels (this is recommended by ImageJ).
- Root length measurement and calculation: We use ImageJ version 1.46r with NeuronJ plugin (http://imagej.nih.gov/ij). Figure 1 shows a photo of a plate, and how root length is measured with imageJ. The scale (1 cm using the graph paper) is measured (Figure 1A), and then with NeuronJ plugin the primary and lateral roots are traced (pink trace, Figure 1B). Values (in cm) are transferred in an Excel file. Total root length is calculated by adding primary and lateral root lengths (see detailed protocol in Figure 1 legend).
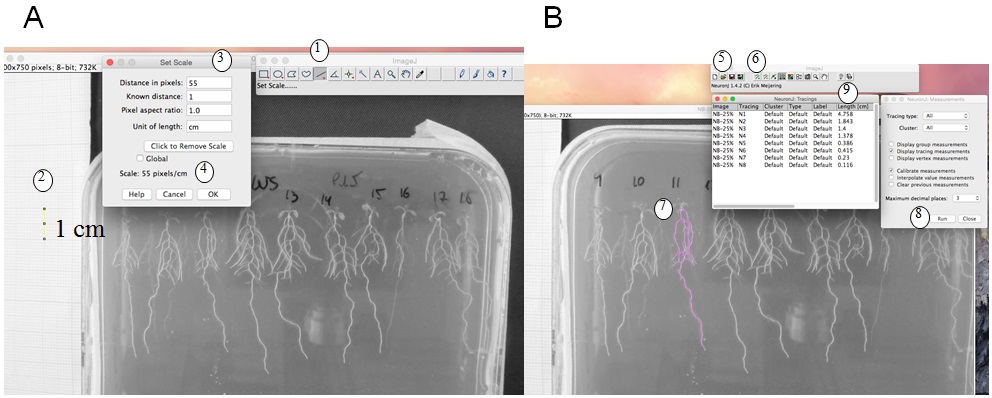
Figure 1. Root length measurement. A. Set the scale: with ImageJ, trace a 1 cm scale on the graph paper (1, 2) then ‘set scale’ (3). This gives pixels/cm (4). B. Measurement of the root length: with NeuronJ plugin, load the image file (5) then set scale (as defined in A). Add tracings (6) by drawing the along the roots (pink trace, 7) then measure tracings (RUN, 8). Results of root length in cm (9) can be copied to an excel file. A and B are screenshots of the process.
- When plating, space the seeds in order to maintain root systems independent for every single plantlet; this will help when measuring the length of the root system (see below). 6 to 10 seeds are sown per plate depending on the growth medium (respectively with high or low Pi, see below). 2 to 3 plates will be necessary per plant type (genotype or treatment depending on the experiment).
- Incubation
- This step should be performed behind a shield for protection against radiation.
- Before starting a series of experiments, you must check that Pi absorption is linear in your conditions (plant specificity, temperature, light). To do that, a time course is performed between 30 min and 2 h following the protocol as described below. In our conditions, Pi absorption was always linear.
- Prepare 12-well plates, one for each plant treatment. Number the wells: 1 to 12.
- Add 4 ml of incubation medium per well. Place 1 plant per well with tweezers (Figure 2), the roots should be immersed and the leaves outside the well. To avoid excessive dehydration of the young plantlets during the incubation, carefully put a cover on the plate avoiding the immersion of the leaves in the solution (Figure 2B). Incubate for 2 h at room temperature (22-24 °C) under white light (150-180 μE m-2 s-1). Discard plants that are fully immersed in the liquid, if any.
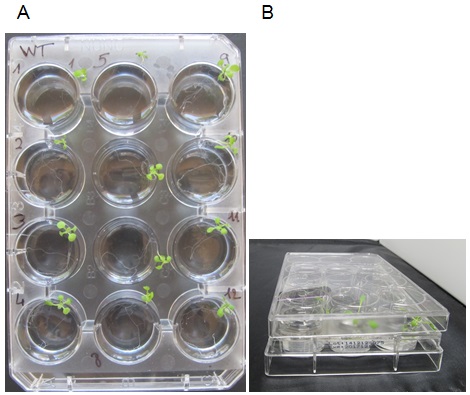
Figure 2. Incubation experiment. A. Photo of a 12-well plate with plants numbered 1 to 12 in the incubation medium. The cover plate has been removed for clarity. Please note that rosettes are out of the medium and roots are fully immersed. B. Photo of the plate and the cover (side view) showing how the cover is placed onto the 12-well plate. - In order to evaluate Pi adsorption on the root (mechanical or chemical adsorption can occur on cells but in that case, Pi does not enter inside the root cells), blank samples are treated as follows: The plant root is dipped in the incubation medium for 2 sec (use tweezers) and directly transferred in the desorption medium for 2 h (see below for the desorption procedure).
- This step should be performed behind a shield for protection against radiation.
- Desorption (see Video 1)Video 1. Desorption
- Prepare new 12-well plates, with 3 ml of cold desorption medium per well. Place the plates on ice.
- Rinse the incubated plants in water (1-2 sec).
- With the help of tweezers, transfer the plants (in the same order you put them in the incubation solution, 1 plant/well) with both leaves and roots immersed in the desorption medium for 2 h on ice.
- Transfer each plant in a counting vial (use tweezers). Alternatively, separate the rosette and roots with a sharp razor and put each part of the plant in a separate vial. It is convenient to put the vials in the counting racks as soon as you harvest the plants (in order to avoid mismatches).
- Prepare new 12-well plates, with 3 ml of cold desorption medium per well. Place the plates on ice.
- Dry the plants in an oven at 50 °C overnight
- Radioactivity measurement
- Under a chemical hood, add 2 ml of scintillation cocktail and tighten the top on each counting vial. Then place the vials in racks adapted to the counter.
- With a beta counter, count 33P in each sample (Cs) and in blanks (Cb is a mean of the blank replicates) and also in the incubation medium (C10, 10 μl of incubation medium are placed in a counting vial and 2 ml scintillation cocktail are added). In the scintillation cocktail, beta radiations are transformed in photons that are detected by the beta counter. The measurement is in cpm (count per min).
- Data analysis:
The amount of PO4 absorbed per hour per root cm (V in nmol/h/cm) is calculated as follows:
V = (Cs-Cb) * (S *10-3)* B / (T * Lroot * C10)
Cs: Radioactivity in the sample (cpm)
Cb: Mean value of radioactivity in the blanks (cpm)
S: Pi concentration in the incubation medium (S=50 μM Pi).
S*10-3=0.05: Pi content (nmol/μl) in the incubation medium (S=50 μM).
C10: Radioactivity in 10 μl of the incubation solution (cpm). C10/(S*10-2) is the specific activity of 33P in the incubation medium
B=10: Volume of the incubation solution for measurement of radioactivity before incubation of the plants (10 μl is convenient).
T=2: Incubation is 2 h
Lroot: Root length (cm)
In addition, using the same equation as above, but omitting Lroot, you can calculate Pi absorption per plant (Cs corresponds to 33P in the whole plant), per rosette tissue or root tissue (Cs corresponds to 33P in the leaves or the root system, respectively).
It is also possible to calculate the ratio of 33P (Cs in leaves in cpm) measured in the leaves compared to total 33P absorbed by the whole plant [Cs in leaves + Cs in roots (cpm)].
- Under a chemical hood, add 2 ml of scintillation cocktail and tighten the top on each counting vial. Then place the vials in racks adapted to the counter.
Representative data
Pi uptake per hour and per root cm for plants grown on low or high Pi (Table 1). Two genotypes are presented (WT and mutant) in order to show the range of variation of Pi uptake between plants and the distribution of Pi in the plant.
Table 1. Pi uptake and distribution in WT and mutant plantlets grown on low or high Pi. Pi uptake capacity is expressed as nmol Pi per hour per root cm (mean±SD). Pi distribution is calculated from the ratio of 33P in leaves compared to 33P in the whole plant.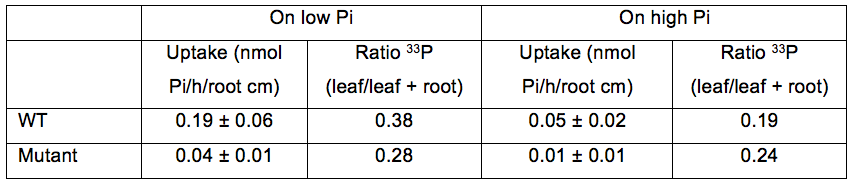
Recipes
- MS/10 medium
10x time diluted Murashige and Skoog medium containing:
0.15 mM MgSO4
2.1 mM NH4NO3
1.9 mM KNO3
0.5 (high Pi) or 0.005 (low Pi) mM NaH2PO4
0.34 mM CaCl2
0.5 μM KI
10 μM FeCl2
10 μM H3BO3
10 μM MnSO4
3 μM ZnSO4
0.1 μM CuSO4
0.1 μM CoCl2
1 μM Na2MoO4
5.9 μM thiamine
4.9 μM pyridoxine
8.1 μM nicotinic acid
55 μM inositol
3.4 mM MES
0.5% sucrose and 0.8% agar at pH 5.7 - Stock solution
0.1 mM CaCl2 in 5 mM MES, adjusted at pH 5.5
For 1 L (water): 0.976 g MES + 0.0147 g CaCl2, adjust pH to 5.5 with 10 N NaOH - 1 M KH2PO4
For 1 L (water): 136 g KH2PO4 - Incubation medium (4 ml/plant are necessary)
50 µM KH2PO4 in stock solution: 50 µl 1 M KH2PO4 in 1 L stock solution
Add 5,550 Bq 33P/ml (0.15 µCi 33P/ml) - Desorption medium (3 ml/plant are necessary)
1 mM KH2PO4 in stock solution: 1 ml 1 M KH2PO4 in 1 L stock solution
Acknowledgments
This protocol was adapted from the previously published studies: Narang et al. (2000) and Misson et al. (2004) and it was performed by Ayadi et al. (2015). This work was supported by the Commissariat à l’Energie Atomique et aux Energies Alternatives.
References
- Ayadi, A., David, P., Arrighi, J. F., Chiarenza, S., Thibaud, M. C., Nussaume, L. and Marin, E. (2015). Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol 167(4): 1511-1526.
- Misson, J., Thibaud, M. C., Bechtold, N., Raghothama, K. and Nussaume, L. (2004). Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol Biol 55(5): 727-741.
- Narang, R. A., Bruene, A. and Altmann, T. (2000). Analysis of phosphate acquisition efficiency in different Arabidopsis accessions. Plant Physiol 124(4): 1786-1799.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Thibaud, M. and Marin, E. (2016). Measurement of 33P-PO4 Absorption Capacity and Root-to-leaf Transfer in Arabidopsis. Bio-protocol 6(5): e1741. DOI: 10.21769/BioProtoc.1741.
- Ayadi, A., David, P., Arrighi, J. F., Chiarenza, S., Thibaud, M. C., Nussaume, L. and Marin, E. (2015). Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol 167(4): 1511-1526.
Category
Plant Science > Plant physiology > Nutrition
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










