- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Virus-based MicroRNA Silencing
Published: Vol 6, Iss 2, Jan 20, 2016 DOI: 10.21769/BioProtoc.1714 Views: 11321
Reviewed by: Feng LiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Extraction of Small RNA and qPCR Validation of miRNAs in Vigna mungo
Sujay Paul [...] Amita Pal
Mar 5, 2015 11473 Views

In vitro Reconstitution Assay of miRNA Biogenesis by Arabidopsis DCL1
Tian Wang [...] Xiuren Zhang
Apr 20, 2015 10877 Views

Effective Gene Silencing in Plants by Synthetic Trans-Acting siRNAs Derived From Minimal Precursors
Adriana E. Cisneros [...] Alberto Carbonell
Oct 20, 2025 1813 Views
Abstract
Virus-based microRNA silencing (VbMS) is a viable and prompt method to screen and characterize the function of microRNAs (miRNAs) in plants. The Tobacco rattle virus (TRV)-based VbMS method was originally developed by the Yule Liu's group (Sha et al., 2014) using miRNA target mimic (TM) methodology. Here, we describe the TRV-based VbMS method for silencing endogenous miRNA in Nicotiana benthamiana and tomato via Agrobacterium infiltrations. For each assay, Agrobacterium cultures containing pTRV1 and specific pTRV2e derivative harboring TM fragments are mixed and infiltrated into plant tissues. Generally within 3 weeks, the target miRNAs gene will be silenced and the newly developed tissues will exhibit corresponding phenotypes.
Keywords: MicroRNAMaterials and Reagents
- Centrifuge tubes
- Sterile 1 ml syringe (needle removed)
- Sterile bacterial culture tubes
- Plant materials
Nicotiana benthamiana, tomato (cultivar Moneymaker)
Note: seeds can be obtained from Yule Liu’s lab. - Bacteria strains
- Escherichia coli: DH5α, ccdb survival (Thermo Fisher Scientific, InvitrogenTM, catalog number: A10460 )
- Agrobacterium tumefaciens: GV3101, GV2260 (alternative to GV3101)
Note: All strains can be obtained from Yule Liu’s lab.
- Escherichia coli: DH5α, ccdb survival (Thermo Fisher Scientific, InvitrogenTM, catalog number: A10460 )
- Plasmids
- pTRV1 (Dong et al., 2007): a T-DNA vector containing 2 x 35 s promoter, Nos terminator and full cDNA of TRV RNA1 (from Ppk20 strain).
- pTRV2e (Sha et al., 2014): a T-DNA vector containing 2 x 35 s promoter, Nos terminator and cDNA clone of TRV RNA2, of which the sub-genomic promoter of coat protein from Pea early brown virus (PEBV) (Wang et al., 1997) and a ligation independent cloning (LIC) cassette are inserted immediately downstream of the TRV CP gene.
- pTRV2e-GFP: GFP gene was inserted at LIC cassette into pTRV2e. This construct can be used in a control assay to show successful exogenous expression.
- The pTRV1 (Arabidopsis, ABRC, catalog number: CD3-1039 ) and pTRV2e (Arabidopsis, ABRC, catalog number: CD3-1866 ) vectors can be ordered at the Arabidopsis Biological Resource Center (ABRC, http://www.arabidopsis.org/).
- pTRV1 (Dong et al., 2007): a T-DNA vector containing 2 x 35 s promoter, Nos terminator and full cDNA of TRV RNA1 (from Ppk20 strain).
- Culture Media
- Liquid Luria-Bertani (LB) medium
Solid LB medium plate with 1.5% agar
Note: LB medium is autoclaved at 120 °C for 20 min before appropriate antibiotics are added.
- Liquid Luria-Bertani (LB) medium
- Antibiotics
- Kanamycin (Sangon Biotech, USP Grade)
- Rifampicin (Sangon Biotech, USP Grade)
- Gentamicin (Sangon Biotech, USP Grade)
- Kanamycin (Sangon Biotech, USP Grade)
- PCR reagents
- EasyTaq DNA polymerase (Beijing TransGen Biotech, catalog number: AP112 )
- EasyPfu DNA polymerase (Beijing TransGen Biotech, catalog number: AP211 )
- dNTP Mix (Roche Diagnostics, catalog number: 04729706103 )
- EasyTaq DNA polymerase (Beijing TransGen Biotech, catalog number: AP112 )
- Infiltration reagents
- Dimethyl sulfoxide, DMSO (AMRESCO, ACS grade)
- MgCl2 (Beijing Chemical Works, Analytical pure) (see Recipes)
- 2-(N-Morpholino) ethanesulfonic acid, MES (AMRESCO, Regent Grade) (see Recipes)
- Acetosyringone (3, 5-Dimethoxy-4-hydroxyacetophenone) (AS) [Sigma-Aldrich, Purity (HPLC)] (see Recipes)
- Infiltration buffer (see Recipes)
- Dimethyl sulfoxide, DMSO (AMRESCO, ACS grade)
Equipment
- Plant growth chamber (24 °C, 16 h/8 h light/dark photoperiod, 40-80% humidity)
- Centrifuge
- PCR instrument
- 37 °C and 28 °C incubators with shaking
Procedure
- TMs designing.
- The TM molecules were designed empirically by adding 3-4 nucleotides into the complementary sequences between sites opposite to the 10th and 11th nt of the targeted miRNA.
Keep the other position with base-paring to the miRNA.
Note: Figure 1 shows an example for TM design.
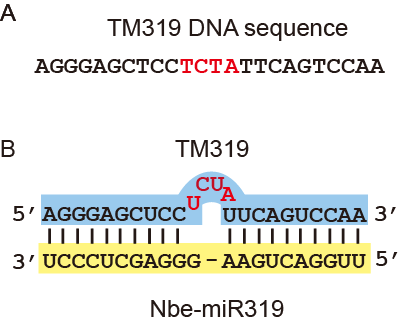
Figure 1. Example for TM design. A. DNA sequence of TM for miRNA319 (TM319). B. Base paring of TM319 and Nbe-miR319.Red letters are nucleotides inserted in to the complementary sequences between sites opposite to the 10th and 11th nt (denoted by “-“). - The TM molecules were designed empirically by adding 3-4 nucleotides into the complementary sequences between sites opposite to the 10th and 11th nt of the targeted miRNA.
- Cloning.
- The miRNA TM fragment was inserted into pTRV2e at LIC cassette as described in the previous study (Sha et al., 2014).
- Correct constructs were screened by PCR and must be confirmed by DNA sequencing.
- The miRNA TM fragment was inserted into pTRV2e at LIC cassette as described in the previous study (Sha et al., 2014).
- Plasmid extraction.
- Correct clones were grown in 2~5 ml liquid LB medium (containing 50 µg/ml Kanamycin) at 37 °C, with shaking at 200 rpm for 16 h.
- Collect the bacteria and extract plasmids using Alkaline Lysis Method (Sambrook, 2001).
- Correct clones were grown in 2~5 ml liquid LB medium (containing 50 µg/ml Kanamycin) at 37 °C, with shaking at 200 rpm for 16 h.
- Agrobacterium transformation.
- Transform pTRV1, pTRV2e or its derivatives into Agrobacterium strain GV3101 (or GV2260) respectively.
- Grown for 2 days on solid LB media (containing 50 µg/ml Kanamycin, 50 µg/ml Rifampicin) at 28 °C.
- Confirm that the Agrobacteria contain desired plasmid by PCR using specific primers and streak the correct transformants on LB plate.
- Transform pTRV1, pTRV2e or its derivatives into Agrobacterium strain GV3101 (or GV2260) respectively.
- Preparation of Agro-infiltrates.
- Grow correct transformants containing pTRV1, pTRV2e or pTRV2e derivatives (Sha et al., 2014, Figure 1) in 5 ml liquid LB media respectively (containing 50 µg/ml Kanamycin, 50 µg/ml Rifampicin) in 28 °C incubator shaking at 200 rpm overnight.
- Collect the culture media and adjust each Agrobacterium culture to OD600=1.0. Mix equal volume of Agrobacterium culture (OD600=1.0) of pTRV1 and that of pTRV2e or pTRV2e derivatives together.
- Pellet the mixed agrobacteria by centrifuging at 3,000 x g for 5 min at room temperature.
- Discard the supernatant and re-suspend the pellet with equal volume of infiltration buffer (to keep OD600 ≈1.0).
- Incubate the re-suspended agrobacteria at room temperature for 2.5-6 h.
Note: Inoculating agrobacteria into media for culturing should be done on clean bench and all equipment used needs to be sterile.
- Grow correct transformants containing pTRV1, pTRV2e or pTRV2e derivatives (Sha et al., 2014, Figure 1) in 5 ml liquid LB media respectively (containing 50 µg/ml Kanamycin, 50 µg/ml Rifampicin) in 28 °C incubator shaking at 200 rpm overnight.
- Plant infiltration.
- Select 6-leaf-stage plants for VbMS assay. Infiltrate the re-suspended agrobacteria into the abaxial side of 3-4 expanded leaves (avoid the midvein) with 1 ml needless syringe.
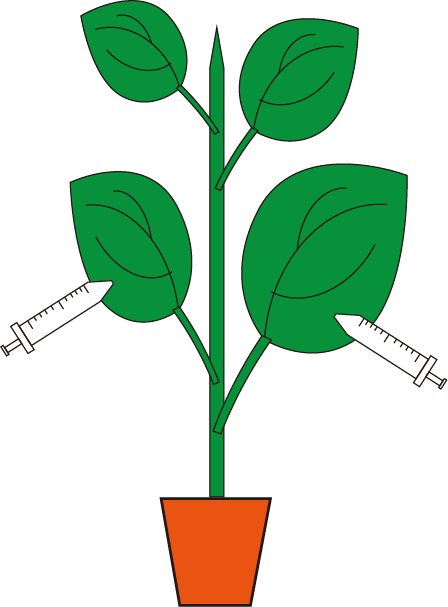
Figure 2. Schematic diagram of Agrobacterium infiltration - Plant growth and evaluation of miRNA silencing effects.
- Grow the infiltrated plants at 24 °C with a 16 h/8 h light/dark photoperiod and the light intensity is 200 μmol m-2 s-1.
In 2-3 weeks post inoculation the target miRNA will be silenced at the whole plant level. - The newly developed organs will show phenotypes of strong silencing of the corresponding miRNA. These tissues can be used for appropriate experiments.
Note: Figure 2 shows a schematic overview of infiltration procedure. Each leaf is often injected at 2-4 sites throughout the leaf lamina, each injection site has a diameter >1 cm.
- Grow the infiltrated plants at 24 °C with a 16 h/8 h light/dark photoperiod and the light intensity is 200 μmol m-2 s-1.
Recipes
- 1 M MgCl2
20.33 g MgCl2.6H2O dissolved in 100 ml ddH2O, autoclaved by 120 °C, 20 min, stored at 4 °C. - 1 M MES
21.325 g MES dissolved in 100 ml ddH2O, sterilized via filtration through 0.22 µm membrane, stored at room temperature. - 200 mM acetosyringone (AS)
0.3924 g AS dissolved in 10 ml DMSO, stored at -20 °C as 1 ml aliquots. - Infiltration buffer (10 mM MgCl2, 10 mM MES, 200 µM AS)
Add 1 ml MgCl2 (1 M), 1 ml MES (1 M), 100 μl AS (200 mM), add ddH2O to 100 ml.
References
- Liu, Y., Schiff, M., Marathe, R. and Dinesh-Kumar, S. P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J 30(4): 415-429.
- Sambrook, J. (2001). Molecular cloning: A laboratory manual, third edition. Cold Spring Harbor Laboratory Press.
- Sha, A., Zhao, J., Yin, K., Tang, Y., Wang, Y., Wei, X., Hong, Y. and Liu, Y. (2014). Virus-based microRNA silencing in plants. Plant Physiol 164(1): 36-47.
- Wang, D., MacFarlane, S. A. and Maule, A. J. (1997). Viral determinants of pea early browning virus seed transmission in pea. Virology 234(1): 112-117.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Zhao, J. and Liu, Y. (2016). Virus-based MicroRNA Silencing. Bio-protocol 6(2): e1714. DOI: 10.21769/BioProtoc.1714.
-
Sha, A., Zhao, J., Yin, K., Tang, Y., Wang, Y., Wei, X., Hong, Y. and Liu, Y. (2014). Virus-based microRNA silencing in plants. Plant Physiol 164(1): 36-47.
Category
Plant Science > Plant molecular biology > RNA > RNA interference
Molecular Biology > RNA > RNA interference
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











