- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Analysis of Starch Synthase Activities in Wheat Grains using Native-PAGE
Published: Vol 6, Iss 2, Jan 20, 2016 DOI: 10.21769/BioProtoc.1713 Views: 11397
Reviewed by: Samik BhattacharyaSaminathan ThangasamyCindy Ast

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

New Approach to Detect and Isolate Rhamnogalacturonan-II in Arabidopsis thaliana Seed Mucilage
Dayan Sanhueza and Susana Saez-Aguayo
Sep 5, 2025 1248 Views

CAPS-Based SNP Genotyping for Nitrogen-Response Phenotypes in Maize Hybrids
Jannis Jacobs [...] Peter K. Lundquist
Dec 20, 2025 550 Views

Detailed Method for the Purification of Rhamnogalacturonan-I (RG-I) in Arabidopsis thaliana
Liang Zhang [...] Breeanna R. Urbanowicz
Feb 5, 2026 144 Views
Abstract
Starch synthases are one class of key enzymes involving in the synthesis of cereal starch, which transfer glucose from ADP-glucose to the non-reducing end of pre-existing α-(1-4)-liked glucosyl chains of amylopectin. This protocol is highly reproducible for assaying activities for starch synthase I and IIIa in wheat and barley endosperm at qualitative level and quantitative level. The protocol includes separating proteins isolated from developing endosperm with native-PAGE containing glycogen from oyster, incubating protein gels with ADP-glucose solution, and staining gels with iodine solution. The method allows researchers to compare the levels or changes of starch synthase activities.
Keywords: Starch synthaseMaterials and Reagents
- Glad-wrap (Microwave safe, Capri)
- Kimwipe (Kimtech Science* Kimwipes delicate task wipers) (Kimberly-Clark, catalog number: 34133 )
- Gel-cassette (size of 100 mm x 100 mm with 1 mm gap) (Invitrogen, catalog number: NC2010 )
Note: Currently, it is “Thermo Fisher Scientific, Novex™, catalog number: NC2010”. - 15 ml blue cap Falcon tube (sterile) (Thermo Fisher Scientific)
- Epipestle (bioWORLD, catalog number: 42741000-1 )
- 96 well UV microplate (Flat bottom) (Thermo Fisher Scientific, catalog number: 8404 )
- Cuvette (plastic) (SARSTEDT AG & Co, catalog number: 67.746 )
- MilliQ water
- 70% ethanol (Chem Supply, catalog number: 64-17-5 )
- 2% agarose (Progen, catalog number: 200-0011 )
- Tris (2-Amino-2-hydroxymethyl-propane-1, 3-diol) (VWR International, catalog number: 103157P )
- Glycogen from oyster (Sigma-Aldrich, catalog number: G8751 )
- Acrylamide (40% Acrylamide/Bis Solution, 37.5:1) (Bio-Rad Laboratories, catalog number: 161-0148 )
- TEMED (Tetramethylethylenediamine) (AMRESCO, catalog number: 0761-25 ML )
- APS (Ammonia persulfate) (Sigma-Aldrich, catalog number: A-7460 )
- Coomassie Plus Protein Assay Reagent (Thermo Fisher Scientific, catalog number: 1856210)
- Bovine serum albumin (BSA) (Freeze dried, Reagent Grade, pH 7) (Moregate Biotech)
- HCl (Ajax Finechem Pty, catalog number: 1367-2.5 L )
- Ammonium Sulfate [(NH4)2SO4] (AR) (Chem Supply, catalog number: AA014-500 G )
- Magnesium Chloride Hexahydrate (MgCl2) (AR) (Chem Supply, catalog number: MA029-500 G )
- β-mercaptoethanol (Sigma-Aldrich, catalog number: M-7154 )
- Adenosine-5’-diphosphoglucose disodium salt (ADPG) (Sigma-Aldrich, catalog number: A0627-250 mg )
- Protease inhibitor cocktail for plant cell and tissue extracts (Sigma-Aldrich, catalog number: P9599 )
- DL-Dithiothreitol (DTT) (Sigma-Aldrich, catalog number: D-9779 )
- Glycine (Chem Supply, catalog number: GA007-500 G )
- Bromophenol blue (Sigma-Aldrich, catalog number: B0126-25 G )
- Glycerol (AR) (Chem Supply, catalog number: GA010-2.5 L )
- Extraction buffer (see Recipes)
- 1x electrophoresis buffer (see Recipes)
Equipment
- Gel tank (XCell SureLock) (Invitrogen, catalog number: EI0001 )
Note: Currently, it is “Thermo Fisher Scientific, NovexTM, catalog number: EI0001”. - Pipette (Thermo Fisher Scientific, model: Finnpipette F1 )
- Spectrophotometer (Agilent Technologies, model: Cary 300 Bio UV-Visible Spectrophotometer )
- Microplate reader (BMG LABTECH GmbH, model: FLUOstar Omega Microplate Reader )
Procedure
- Prepare 8% Native-PAGE gels
- Preparing gel cassettes
Both commercial cassettes and self assembled cassettes with glass plates can be used. If not using commercial cassettes, glass plates, 1 mm spacers and combs need to be thoroughly cleaned with MilliQ water and then with 70% ethanol (use of 1 mm gels gives better resolution than 1.5 mm gels). Glass plates need to be secured with clamps - don’t take clamps past inside of spacers. The bottom of plates is sealed by standing plates in molten 1 ml of 2% agarose in 0.3 M Tris/HCl, pH 8.8 poured onto Glad-wrap. - Preparing separating gel layer (8% Native-PAGE) as Table 1
- First dissolve 34.5 mg glycogen from oyster in 3.473 ml water (for 1 gel) or 69 mg glycogen in 6.945 ml water (for 2 gels) in a 15 ml blue cap Falcon tube.
- Add 1 M Tris/HCl, pH 8.8 and 40% acrylamide (37.5:1).
- Degas the gel solution for 5 min to remove air using water tap vacuum.
- Add TEMED and 10% APS.
Table 1. Chemicals and solutions for separating gels1 gel (7.5 ml) 2 gels (15 ml) Glycogen from oyster 34.5 mg 69 mg H2O 3.473 ml 6.945 ml 1 M Tris/HCl, pH 8.8 2.475 ml 4.95 ml 40% acrylamide (37.5:1) 1.5 ml 3 ml Add items below last for polymerizing gel TEMED 7.5 μl 15 μl 10% APS 45 μl 90 μl
Notes:- *Tips and tubes used for acrylamide solution need to be discarded in the acrylamide waste bin for toxic waste management.
- 10% APS: Make 500 μl solution and aliquot to 65 μl per tube and store at -20 °C. Take one tube out of -20 °C freezer each time to use. The same tube should only be used for 1 week after thawing and storing at 4 °C. Discard the tube more than 1 week after thawing.
- *Tips and tubes used for acrylamide solution need to be discarded in the acrylamide waste bin for toxic waste management.
- Pour the gel mix into the cassette to 1-1.5 cm below the bottom of the comb using 1 ml pipette. Add 0.5 ml of isopropanol on the top of the gel mix inside of the cassette to level the top of the gel and to aid in the gel polymerisation. Leave the gel mix for polymerisation for ~30 min at room temperature.
- Before adding stacking gel layer, pour off isopropanol and wash at least twice with MilliQ H2O. Carefully wipe MilliQ H2O between the two plates with a Kimwipe to remove as much excess MilliQ H2O as possible without touching the gel.
- First dissolve 34.5 mg glycogen from oyster in 3.473 ml water (for 1 gel) or 69 mg glycogen in 6.945 ml water (for 2 gels) in a 15 ml blue cap Falcon tube.
- Preparing stacking gel layer
- Make stacking gel mix in a 15 ml blue cap Falcon tube as Table 2.
Table 2. Solutions for stacking gels1 gel (2.5 ml) 2 gels (5 ml) 0.5 M Tris/HCl (pH 6.8) 650 μl 1.3 ml 40% acrylamide (37.5:1) 285 μl 570 μl H2O 1.5475 ml 3.095 ml Add items below last for polymerizing gel TEMED 2.5 μl 5 μl 10% APS 15 μl 30 μl - Pour the stacking gel into the cassette and add a comb. If pouring more than one gel, add the comb to the stacking gel of the first gel poured before pouring the second gel. Leave the stacking gel mix for another ~30 min for polymerization.
- If it has been planned to run the gel the next day, the gel needs to be covered with Glad-wrap and stored at 4 °C.
- Make stacking gel mix in a 15 ml blue cap Falcon tube as Table 2.
- Preparing gel cassettes
- Prepare protein samples
- Protein extraction
The following steps for protein extraction need to be processed on ice or 4 °C to avoid protein degradation.- Take 1-2 developing endosperms (12 to 15 days after anthesis) and place in an Eppendorf tube, and then weigh.
- Add 100 μl of extraction buffer (see Recipes) per 100 mg tissue.
- Grind seeds to a pulp using an epipestle, spin at 14,000 x g for 15 min at 4 °C.
- Transfer supernatant into a new Eppendorf tube.
- If sediment materials are mixed with supernatant, spin for 5-10 min at 4 °C again and transfer supernatant into another new Eppendorf tube.
- Take 1-2 developing endosperms (12 to 15 days after anthesis) and place in an Eppendorf tube, and then weigh.
- Protein quantification
- Prepare a protein concentration standard curve
- Prepare BSA solution (0.25 mg/ml) by diluting BSA solution (1 mg/ml) in a ratio of 1:4 with MilliQ H2O.
- Construct a standard curve for protein concentration using BSA (0.25 mg/ml) as in Table 3 for 1 ml cuvette or Table 4 for 96 well UV microplate.
Table 3. Preparation of protein standards for 1 ml cuvetteProtein concentration (μg/1,100 μl) BSA volume (μl) H2O (μl) Coommassie plus protein assay reagent (μl) 0 0 100 900 5 20 80 900 10 40 60 900 15 60 40 900 20 80 20 900 25 100 0 900
Table 4. Preparation of protein standards for 96 well UV microplateProtein concentration (μg/220 μl) BSA volume (μl) H2O (μl) Coommassie plus protein assay reagent (μl) 0 0 20 180 5 4 16 180 10 8 12 180 15 12 8 180 20 16 4 180 25 20 0 180
- Prepare BSA solution (0.25 mg/ml) by diluting BSA solution (1 mg/ml) in a ratio of 1:4 with MilliQ H2O.
- Preparation of samples
For 1 ml cuvette, add 3 μl of sample to 97 μl H2O and 900 μl of Coomassie Plus Protein Assay Reagent.
For 96 well plate, add 1 μl (or less) of sample to 19 μl H2O (or more) and 180 μl of Coomassie Plus Protein Assay Reagent. - Measure the protein absorbance with a spectrophotometer for cuvettes or a microplate reader for 96 well UV microplate
- Assay protein standards and samples on a spectrophotometer or a microplate reader.
- Read absorbance at 595 nm.
- Match absorbance against a standard curve. Divide μg in sample by number of μl added, in this case 3, to give final concentration in μg/μl.
- Check that color falls within color range of standards. If not, reduce or increase amount of sample accordingly.
- Store protein samples by freezing samples in liquid nitrogen and storing at -80 °C.
- Assay protein standards and samples on a spectrophotometer or a microplate reader.
- Prepare a protein concentration standard curve
- Read absorbance for protein standards and samples at the same time.
- Construct protein standard curve in Excel as Figure 1:
- Construct scatter chart.
- Select data in the Excel sheet, click “Insert” tab, then “Charts” tab, click “Scatter” icon, and add “Trendline”.
- Under the Type tab, select “Linear”.
- Check “Set Intercept = 0” and check “Display equation on chart” and “Display R-squared value on chart”.
- Use the equation to determine protein content of samples (Don’t forget to divide by the number of μl of sample added to give concentration in μg/μl).

Figure 1. A standard curve for measuring protein concentration using BSA as protein standard. The X-axis is the protein concentration and the Y-axis is the absorbance measured at 595 nm.
- Construct scatter chart.
- Protein extraction
- Gel electrophoresis
- Assemble gels.
- Pour 800 ml 1x electrophoresis buffer into gel tank.
- Add the loading buffer to protein sample solution on ice.
Perform following steps in fume hood:
To each 100 μl of protein sample solution, add:
10 μl 0.5 M Tris-HCl (pH 6.8)
10 μl β-mercaptoethanol
5 μl 0.6% bromophenol blue in 80% glycerol - Re-calculate concentration to allow for addition of above denaturing buffer to proteins. New final concentration= (initial volume/final volume) x initial concentration.
- Load 100 μg total protein for each sample in each well.
- If space permits, leave a lane between ladder and samples.
- Run gels at a maximum voltage at 100 V (or a maximum ampere at 15 mA per gel).
- Running gels until the bromophenol blue dye reaches the bottom of the gel.
- Assemble gels.
- Assay starch synthase activity
- Prepare starch synthase assay buffer as Table 5.
Table 5. Starch synthase assay buffer10 ml 15 ml Note 10x Tris-Glycine
(10x running buffer)1 ml 1.5 ml 2 M (NH4)2SO4 667 μl 1 ml 2 M MgCl2 37μl 55 μl BSA (dried) 6.7 mg (0.0067 g) 10 mg (0.010 g) β-mercaptoethanol 47 μl 70 μl 360 mM ADPG (-20 °C) 34 μl 51 μl Add last as it goes off quickly H2O 8.22 ml 12.32 ml
2 M MgCl2 2 M (NH4)2SO4 MW=203.3 g
2 M= 4.066 g/10 mlMW= 132.13 g
2 M= 2.643 g/10 ml
Note: 360 mM ADPG needs to be kept as 34 μl aliquot and kept at -20 °C. Take out one tube for each gel from freezer before use. - Incubate the gel(s) with starch synthase assay buffer overnight at room temperature (~22 °C).
- Wash gel(s) two times with MilliQ H2O.
- Stain gel(s) with iodine solution (containing 2% KI, 0.2% I2) until brown bands appear as Figure 2, usually about 10 min. Change iodine solution after 10 min if necessary.
- The gels can be kept in iodine solution if needed.
- Prepare starch synthase assay buffer as Table 5.
Representative data
- Representative examples of starch synthase activity results can be expected in barley grain as Figure 2(b) (from Figure 8A, Li et al., 2011) and in wheat grain as Figure 2(c) (From Figure 2, McMaugh et al., 2014).
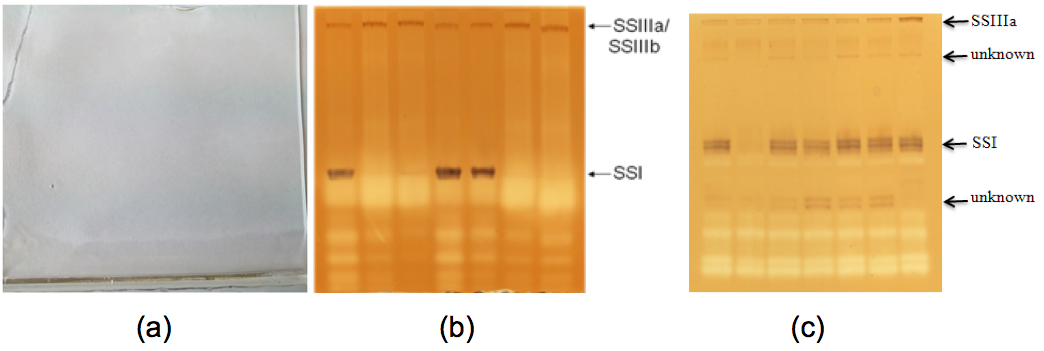
Figure 2. Starch synthase activity of developing endosperm of barley and wheat. (a). A native-PAGE gel before staining with iodine solution. (b). A native-PAGE gel showing starch synthase I and IIIa activities in barley developing endosperm after staining with iodine solution. (c). A native-PAGE gel showing starch synthase I and IIIa activities in wheat developing endosperm after staining with iodine solution. - This protocol is highly reproducible in our hand. The quality of native polyacrylamide gels determines the quality of protein bands. The quality of both TEMED and APS may affect the gel quality and then protein band quality.
Notes
- Glycogen from oyster is the best substrate for native polyacrylamide gel (native-PAGE).
Recipes
- Extraction buffer
Add 20 μl of 1 M potassium phosphate buffer, pH 7.5 (final concentration: 50 mM KPi)
Add 1 μl of 500 mM EDTA, pH 7.5 (final concentration: 5 mM EDTA)
Add 400 μl of 50% glycerol (final concentration: 20% glycerol)
Add 567 μl MilliQ H2O
Add Protease inhibitor cocktail for plant cell and tissue extracts and DTT to buffer as below when ready to extract
Add 2 μl Protease inhibitor cocktail for plant cell and tissue extracts
Add 10 μl of 500 mM DTT (final concentration: 5 mM DTT) - 1x electrophoresis buffer
Add 80 ml 10x running buffer
Add DTT at 0.15 g /L
Make up to 800 ml with distilled water
10x running buffer (10x Tris-glycine Stock Solution)
1.9 M glycine (144 g/L)
250 mM Tris (30.3 g/L)
Acknowledgments
This protocol was modified from previous publication (Abel et al., 1996). The author thanks Behjat Kosar-Hashemi, Emma Anschaw, Steve McMaugh, Sapna Vibhakaran Pillai and Hong Wang for their contributions in optimizing and testing this protocol. The author also thanks CSIRO Plant Industry, CSIRO Food Future Flagship and ACVL Ltd for providing funding resource for testing this protocol during the course of research.
References
- Abel, G. J., Springer, F., Willmitzer, L. and Kossmann, J. (1996). Cloning and functional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato (Solanum tuberosum L.). Plant J 10(6): 981-991.
- Li, Z., Mouille, G., Kosar-Hashemi, B., Rahman, S., Clarke, B., Gale, K. R., Appels, R. and Morell, M. K. (2000). The structure and expression of the wheat starch synthase III gene. Motifs in the expressed gene define the lineage of the starch synthase III gene family. Plant Physiol 123(2): 613-624.
- Li, Z., Li, D., Du, X., Wang, H., Larroque, O., Jenkins, C. L., Jobling, S. A. and Morell, M. K. (2011). The barley amo1 locus is tightly linked to the starch synthase IIIa gene and negatively regulates expression of granule-bound starch synthetic genes. J Exp Bot 62(14): 5217-5231.
- McMaugh, S. J., Thistleton, J. L., Anschaw, E., Luo, J., Konik-Rose, C., Wang, H., Huang, M., Larroque, O., Regina, A., Jobling, S. A., Morell, M. K. and Li, Z. (2014). Suppression of starch synthase I expression affects the granule morphology and granule size and fine structure of starch in wheat endosperm. J Exp Bot 65(8): 2189-2201.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Li, Z. (2016). Analysis of Starch Synthase Activities in Wheat Grains using Native-PAGE. Bio-protocol 6(2): e1713. DOI: 10.21769/BioProtoc.1713.
Category
Plant Science > Plant biochemistry > Carbohydrate
Plant Science > Plant physiology > Nutrition
Biochemistry > Carbohydrate > Polysaccharide
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










