- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro CLE Peptide Bioactivity Assay on Plant Roots
Published: Vol 5, Iss 24, Dec 20, 2015 DOI: 10.21769/BioProtoc.1689 Views: 10493
Reviewed by: Tie LiuFang XuArsalan Daudi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Assessing Metabolite Interactions With Chloroplastic Proteins via the PISA Assay
Anna Karlsson [...] Elton P. Hudson
May 5, 2025 2010 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1772 Views

Quantitative Analysis of the Arabidopsis Leaf Secretory Proteome via TMT-Based Mass Spectrometry
Sakharam Waghmare [...] Rucha Karnik
Nov 20, 2025 2025 Views
Abstract
Plant CLAVATA3/ESR (CLE)-related proteins play diverse roles in plant growth and development including regulating the development of root meristem. The mature functional forms of CLE peptides are typically 12-13 amino acids (aa) in length that are derived from the conserved C-termini of their precursor proteins. Genes encoding small secreted peptides sharing similarity to plant CLE proteins have recently been cloned from plant-parasitic nematodes, pests that infect many important crops. It is demonstrated that exogenous application of synthetic 12-14 aa CLE peptides corresponding to the CLE domain of their precursor proteins can suppress plant root growth. This protocol is to evaluate the bioactivity of CLE peptides originated from plant-parasitic nematodes by measuring the growth of plant roots or the size of root apical meristem (RAM) after CLE peptide treatment. Plants used in the study included Arabidopsis and potato.
Keywords: CLE peptideMaterials and Reagents
- Micropore tape (Thermo Fisher Scientific, catalog number: 19-027-761 )
- Sterile square petri dishes (VWR International, catalog number: 60872-310 )
- Sterile 6-well plates (Greiner Bio-One GmbH, catalog number: 657185 )
- 1.7 ml microcentrifuge tubes (Laboratory Products Sales, catalog number: L211511 )
- 50 ml conical centrifuge tubes
- Microscope slides (Laboratory Products Sales, catalog number: M130400 )
- Sterile steel blades (Staples, catalog number: 111322 )
- Arabidopsis thaliana (Columbia-0) seeds (https://www.arabidopsis.org/abrc/index.jsp)
- Potato true seeds (Solanum tuberosum) (http://www.ars-grin.gov/nr6/)
- Potato plant (Solanum tuberosum cv. Désirée) (http://www.ars-grin.gov/nr6/)
- CLE peptides (> 70% purity) (Selleck Chemicals LLC) (www.selleckchem.com)
- Bleach (6% sodium hypochlorite) (Clorox Company)
- Sterile distilled water
- Agarose (Thermo Fisher Scientific, catalog number: BP160-100 )
- Timentin (PhytoTechnology Laboratories®, catalog number: T869 )
- Thiamine hydrochloride (C12H17ClN4OS.HCl) (Sigma-Aldrich, catalog number: T1270 )
- MS salt (Caisson Laboratories, catalog number: MSP01-50LT )
- Inositol (Thermo Fisher Scientific, catalog number: AC122261000 )
- 2-(4-Morpholino)ethane Sulfonic Acid (MES) (Thermo Fisher Scientific, catalog number: BP300-100 )
- Sodium Phosphate Monobasic Monohydrate (NaH2PO4.H2O) (Thermo Fisher Scientific, catalog number: S369-500 )
- Sucrose (Thermo Fisher Scientific, catalog number: S5-12 )
- Gelrite (Thermo Fisher Scientific, catalog number: CAS 71010-52-1 )
Note: Currently, it is “Sigma-Aldrich, catalog number: CAS 71010-52-1 ”. - Agar (Thermo Fisher Scientific, catalog number: BP1423-500 )
- Propagation medium (1 L) (see Recipes)
- ½ MS medium (1 L) (see Recipes)
- 0.1% agarose (see Recipes)
- Sodium phosphate buffer (see Recipes)
Equipment
- Sterile forceps and scalpel (sterilized by heat treatment using a Bunsen burner)
- 360° Multi-functional tube rotator (Thermo Fisher Scientific, Barnstead Thermolyne, model: Labquake Shaker Rotisserie )
- Growth chamber (Percival Scientific, model: I-66LLVL )
- Biosafety cabinet (NuAire, model: Class II Type A/B3 )
- Olympus BX-50 microscope
- Digital CCD camera (QImaging, model: Retiga EXi )
Software
- MetaMorph imaging analysis software
(http://core.phmtox.msu.edu/Scheduling/ItemDocs/9/MetaMorph_Software_Basic_Analysis_Guide.pdf)
Procedure
- Peptide preparation (performed under sterile conditions)
- Dissolve synthesized CLE peptides in filter-sterilized 50 mM sodium phosphate buffer to make a final stock concentration of 10 mM.
- Aliquot 200 µl of the peptide stock into 1.7 ml microfuge tubes and store them in a -80 °C freezer for later uses.
- Dissolve synthesized CLE peptides in filter-sterilized 50 mM sodium phosphate buffer to make a final stock concentration of 10 mM.
- Peptide assay with plant seeds (performed under sterile conditions)
- Measure out approximately 100 seeds (Arabidopsis or potato) for each peptide treatment and transfer the seeds to a labeled 1.7 ml microcentrifuge tube.
- Add 1 ml 50% bleach to the tube and incubate the seeds in bleach for 15 min (for Arabidopsis seeds) or 5 min (for potato true seeds) on a rotator. During the incubation time, if seeds clump in the tube bottom, resuspend the seeds by gentle shaking and then put the tube back on the rotator.
- Remove the bleach solution and rinse the seeds with 1.5 ml sterile distilled water for three times (5 min each on a rotator) to remove any trace of bleach.
- Remove sterile water after last wash. For Arabidopsis seeds, add 1 ml of sterile 0.1% agarose solution to the tube, resuspend the seeds by gentle shaking and store the tube at 4 °C for 3 d (with light or in darkness) before plating on the plates. For potato true seeds, plate them directly on agar plates.
- Add the peptide stock solution to 30 ml of ½ MS agar medium to achieve desirable peptide concentrations (e.g., 5 µM or 10 µM). After mixing up, pour the medium into a square plate and use the agar plate until the medium solidified. Agar plates containing the same volume of 50 mM sodium phosphate buffer as the added peptide stock will be used as controls.
- Plate approximately 12 seeds on the top of a square plate.
- Place the plates vertically in a growth chamber and culture at 24 °C under a 16-h/8-h light/dark cycle.
- Measure the root length (using a ruler) from the base of the hypocotyl to the tip of the primary root at several time points (e.g., 7 d and 14 d) after plating (Figure 1), or measure the size of the root apical meristem (RAM) of each root tip (Figure 2). Roots of 10 seedlings were measured at each time point.
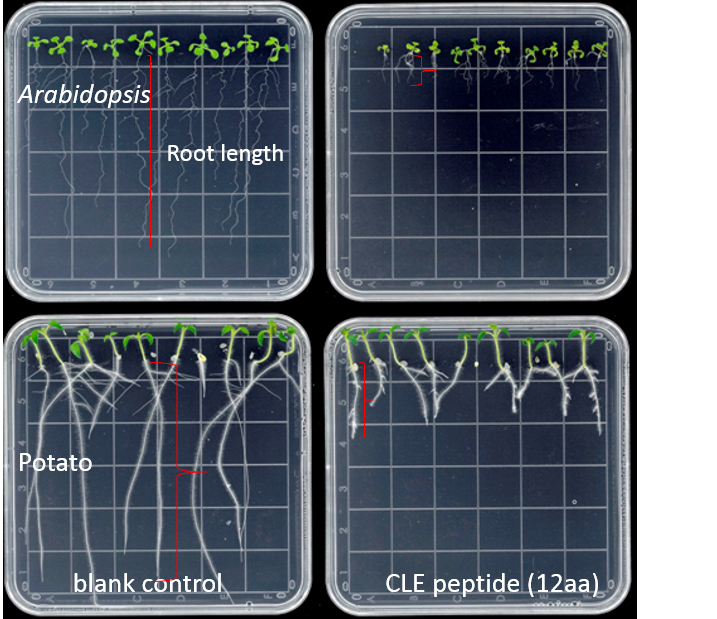
Figure 1. Effects of 10 µM GrCLE1-1 peptide (RVTPGGPDPLHN) on the growth of Arabidopsis and potato roots 10 d after peptide treatment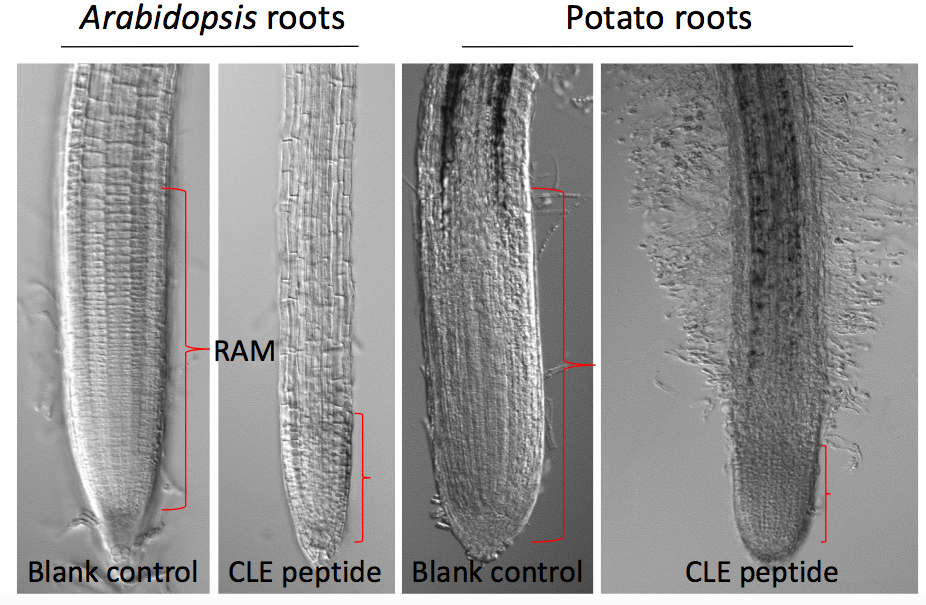
Figure 2. Effects of 10 µM GrCLE1-1 peptide (RVTPGGPDPLHN) on the development of root apical meristem of Arabidopsis and potato roots 14 d after peptide treatment
- Measure out approximately 100 seeds (Arabidopsis or potato) for each peptide treatment and transfer the seeds to a labeled 1.7 ml microcentrifuge tube.
- Peptide assay on tissue-cultured potato plantlets (performed under sterile conditions)
- Plate internode stem segments (approximately 1 cm long) cut using sterile steel blade from in vitro grown potato plantlets into each well of a 6-well plate that contains 6 to 7 ml of propagation medium with certain concentrations of the tested CLE peptide and timentin (100 μg/ml). The top part of the internode segment should be exposed in the air. For the test using internode stem pieces, usually higher peptide concentrations (e.g., 5 μM) need to be used in order to observe an effect on root growth.
- Label plates and seal them with micropore tapes.
- Cultivate plates in a growth chamber at 24 °C under a 16-h/8-h light/dark cycle for two weeks and then measure the RAM size of root tips.
- Plate internode stem segments (approximately 1 cm long) cut using sterile steel blade from in vitro grown potato plantlets into each well of a 6-well plate that contains 6 to 7 ml of propagation medium with certain concentrations of the tested CLE peptide and timentin (100 μg/ml). The top part of the internode segment should be exposed in the air. For the test using internode stem pieces, usually higher peptide concentrations (e.g., 5 μM) need to be used in order to observe an effect on root growth.
- Measurement of the RAM size of root tips
- Use a steel blade to cut off the primary root tips (approximately 2 cm long) from peptide-treated plant seedlings.
- Mount the root tips immediately in water onto a glass slide and examine under an Olympus BX-50 microscope equipped with Nomarski optics.
- Capture the images using a QImaging Retiga EXi digital CCD camera.
- Measure microscopically the root apical meristem region of individual primary roots covering the distance from the position around the quiescent center to the position where elongated root cells immediately follow cytoplasm-dense meristematic cells (Figure 2).
- Analyze the collected data using the MetaMorph imaging analysis software (The manual can be found in the following link: http://core.phmtox.msu.edu/Scheduling/ItemDocs/9/MetaMorph_Software_Basic_Analysis_Guide.pdf).
- Use a steel blade to cut off the primary root tips (approximately 2 cm long) from peptide-treated plant seedlings.
Recipes
- Propagation medium (1 L)
4.3 g MS salt
0.17 g NaH2PO4.H2O
0.10 g inositol
0.4 mg thiamine HCl
30 g sucrose
2.5 g gelrite
Adjust pH to 6.0 with KOH and autoclave (121 °C, 20 min) - ½ MS medium (1 L)
2.15 g MS salt
10 g sucrose
0.5 g MES
15 g agar
Adjust pH to 5.8 with KOH and autoclave (121 °C, 20 min) - 0.1% agarose
0.1 g agarose in 100 ml distilled water
Autoclave and store at room temperature - 50 mM sodium phosphate buffer
44 mM NaH2PO4
6 mM Na2HPO4
Adjust pH to 6.0 with NaOH and filter sterilize
Acknowledgments
This work was supported by funding from USDA-ARS and the USDA-NRI Competitive Grants program.
References
- Chen, S., Lang, P., Chronis, D., Zhang, S., De Jong, W. S., Mitchum, M. G. and Wang, X. (2015). In planta processing and glycosylation of a nematode CLAVATA3/ENDOSPERM SURROUNDING REGION-like effector and its interaction with a host CLAVATA2-like receptor to promote parasitism. Plant Physiol 167(1): 262-272.
- Chronis, D., Chen, S., Lang, P., Tran, T., Thurston, D. and Wang, X. (2014). In vitro nematode infection on potato plant. Bio-protocol 4(1): e1016.
- Fiers, M., Golemiec, E., Xu, J., van der Geest, L., Heidstra, R., Stiekema, W. and Liu, C. M. (2005). The 14-amino acid CLV3, CLE19, and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17(9): 2542-2553.
- Lu, S. W., Chen, S., Wang, J., Yu, H., Chronis, D., Mitchum, M. G. and Wang, X. (2009). Structural and functional diversity of CLAVATA3/ESR (CLE)-like genes from the potato cyst nematode Globodera rostochiensis. Mol Plant Microbe Interact 22(9): 1128-1142.
- Miyawaki, K., Tabata, R. and Sawa, S. (2013). Evolutionarily conserved CLE peptide signaling in plant development, symbiosis, and parasitism. Curr Opin Plant Biol 16(5): 598-606.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Chen, S. and Wang, X. (2015). In vitro CLE Peptide Bioactivity Assay on Plant Roots. Bio-protocol 5(24): e1689. DOI: 10.21769/BioProtoc.1689.
- Chen, S., Lang, P., Chronis, D., Zhang, S., De Jong, W. S., Mitchum, M. G. and Wang, X. (2015). In planta processing and glycosylation of a nematode CLAVATA3/ENDOSPERM SURROUNDING REGION-like effector and its interaction with a host CLAVATA2-like receptor to promote parasitism. Plant Physiol 167(1): 262-272.
Category
Plant Science > Plant biochemistry > Protein
Plant Science > Plant physiology > Plant growth
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











