- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Luminol-based Assay for Detection of Immunity Elicitor-induced Hydrogen Peroxide Production in Arabidopsis thaliana Leaves
Published: Vol 5, Iss 24, Dec 20, 2015 DOI: 10.21769/BioProtoc.1685 Views: 22064
Reviewed by: Michael EnosZhaohui LiuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Rice Ragged Stunt Virus Propagation and Infection on Rice Plants
Chao Zhang [...] Jianguo Wu
Oct 20, 2018 6504 Views

Botrytis cinerea in vivo Inoculation Assays for Early-, Middle- and Late-stage Strawberries
Piao Yang [...] Ye Xia
Oct 20, 2023 2775 Views
Abstract
In Arabidopsis thaliana, one of the very early immune-related responses induced after elicitor perception is the oxidative burst, i.e., reactive oxygen species (ROS) generation including superoxide anion and hydrogen peroxide (H2O2). ROS production plays different roles in a wide range of biotic and abiotic stress responses, including the closure of stomata and the regulation of cell expansion. In particular, elicitor-induced H2O2 is produced mainly by the membrane localized NAD(P)H oxidases RESPIRATORY BURST OXIDASE HOMOLOGUE D and F. In this protocol, we describe a simple and reproducible luminol/peroxidase-based assay to detect and evaluate immunity-related accumulation of H2O2 produced in Arabidopsis leaf discs treated with immunity elicitors, such as oligogalacturonides (OGs), flagellin (flg22) or the elongation factor-thermo-unstable (EF-Tu - elf18). This method is based on the detection of the luminescence released by excited-luminol molecules generated after the horseradish peroxidase (HRP)-catalyzed oxidation of luminol molecules in the presence of H2O2. Levels as well as duration of the luminescence are proportional to the amount of H2O2 produced by elicited leaf discs.
Materials and Reagents
- Petri plates (90 x 15 mm)
- 96-well luminometer plate (Thermo Fisher Scientific, catalog number: 136101 )
- Aluminum foil
- At least two Arabidopsis thaliana (e.g., ecotype Columbia-0) plants for each elicitor treatment
- Luminol (Sigma-Aldrich, catalog number: 123072 )
- Peroxidase from horseradish (Sigma-Aldrich, catalog number: P8125 )
- Sterile distilled water
- Dimethyl sulfoxide (DMSO) (Sigma-Aldrich, catalog number: 41640 )
- Flg22 (QRLSTGSRINSAKDDAAGLQIA) and elf18 (ac-SKEKFERTKPHVNVGTIG)-EZBiolab (http://www.ezbiolab.com/)
- OGs with an average degree of polymerization of 10 to 16 prepared as previously described (Bellincampi et al., 2000)
- 500x luminol stock solution (LSS) (see Recipes)
- 500x horseradish peroxidase stock solution (HPSS) (see Recipes)
- 1 mM flg22 stock solution (see Recipes)
- 1 mM elf18 stock solution (see Recipes)
Equipment
- Arabidopsis growth chamber
- Surgical blade
- Cork borer set (0.125 cm2 area) (Sigma-Aldrich, catalog number: Z165220 )
- Plastic bur block
- Flat tweezer
- GloMax®-Multi+ Detection System with dual injectors (Promega Corporation)
- Microsart® e.jet laboratory vacuum pump (Sartorius AG, catalog number: 16612 )
- Plastic or glass desiccator
Procedure
- Preparation of plant material
Note: At least 12 leaf discs are used for each elicitor treatment. Collect at least 3-4 leaves from each plant considering that, depending on leaf size, 2-3 discs can be obtained. In the case of OG treatment four additional discs are required as a negative control (see below).- Grow Arabidopsis plants in a growth chamber until pre-bolting stage (4-5 weeks) at optimum condition (22 °C, 70% relative humidity, and a photoperiod of 12/12 h light/dark).
- Select and harvest appropriate leaves (third to fifth from the top) from at least two different plants, by cutting them from the petiole with a surgical blade. Gently wash harvested leaves with sterile distilled water (SDW).
Note: Appropriate leaves are defined as leaves from plants that i) do not have different height and age, due to non-synchronous germination; ii) are not wounded; iii) do not show any disease symptoms caused by the presence of soil fungi and/or bacteria. - To produce leaf discs, place the leaf above the bur block with the midrib in the correspondence of one of the holes. Punch the leaf with the cork borer positioned exactly above the hole (see Figure 1A-B).
- Collect and recover leaf discs for 2 h in a Petri dish containing abundant SDW (25 ml) in dark condition, by covering the plate with aluminum foil. Replace the SDW every 30 min (see Figure 1C).
Note: At this point of the procedure the leaf discs do not need to be completely immersed in water. However, it is important that they are well in contact with the aqueous medium to prevent dehydration and to remove secondary metabolites produced upon wounding. - Gently place, by using a flat tweezer, one leaf disc per well in a 96-well luminometer plate each containing 200 µl SDW (see Figure 1D).
- Cover the plate with aluminum foil and incubate in the growth chamber (see step A1 for specifications) overnight.
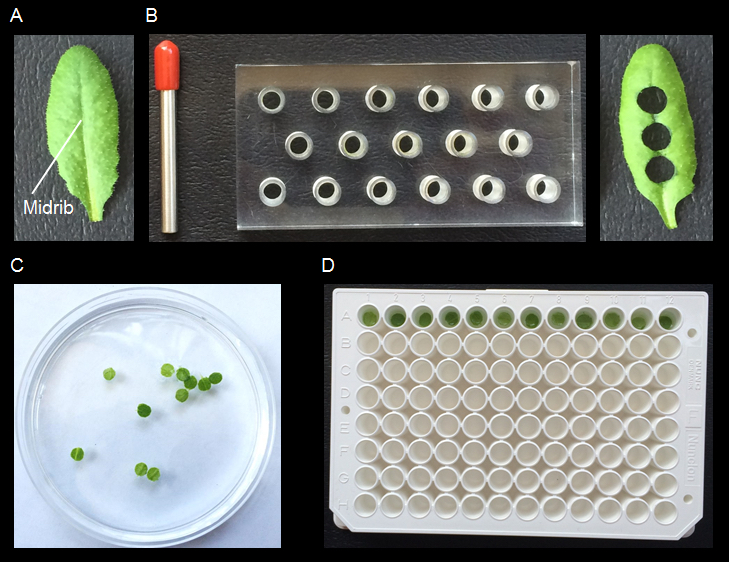
Figure 1. Preparation of plant material. Arabidopsis leaf A is punched at the level of the midrib by using a bur block and a cork borer B. The resulting leaf discs are rinsed with 25 ml of SDW in Petri dish for 2 h C, replacing the SDW every 30 min. The leaf discs are then placed in a 96-well luminometer plate, 1 disc/1 well containing 200 µl SDW D.
- Grow Arabidopsis plants in a growth chamber until pre-bolting stage (4-5 weeks) at optimum condition (22 °C, 70% relative humidity, and a photoperiod of 12/12 h light/dark).
- Detection of flg22- or elf18-triggered H2O2 production
- Prepare stock solutions (500x) both for luminol (LSS) and horseradish peroxidase (HPSS) in SDW. Aliquots of both LSS an HPSS can be stored at -20 °C for further uses.
- Prepare a luminol/peroxidase working solution (1.25x each) in SDW; consider that you will need 200 µl of this solution for each well in the analysis.
Example: For 12 wells mix 6 µl LSS with 6 µl HPSS up to 2,400 µl with SDW. After use, discard the solution. - Prepare 1 mM flg22 or elf18 stock solution (FSS or ESS) in SDW. Stock solution can be aliquoted and stored at -20 °C for further uses.
- Prepare a 5x working solution containing flg22 or elf18 (FWS or EWS) in SDW; consider that you will need 50 µl per well and 1 ml of dead volume to charge the injector. The elicitor final concentration may be comprised between 0.1 µM and 1 µM.
Example: For 12 wells mix 0.8 µl FSS or ESS up to 1600 µl with SDW to obtain 0.5 µM FWS or EWS. After use discard the solution. - Gently replace the 200 µl of water with an equal volume of luminol/peroxidase working solution in each well containing leaf disc.
Critical step: Avoid leaf discs injuries; make sure to completely submerge the leaf discs in the luminol/peroxidase solution prior elicitor injection.
Note: Since leaf discs have hydrophobic surfaces gently use a pipette tip, to fully immerse both sides of the leaf disc in the solution. - Manually place the plate in the GloMax®-Multi+ Detection System (see Figure 2). Charge one of the injectors with the 5x elicitor solution and aliquot 50 µl per well.
- Perform the luminescence detection assay for at least 40 min with a signal integration time of 1 sec.
Note: The indicated time only refers to the reading of the plate, which is done in stationary stage and in automated way by the detection system (for the manufacturer’s instructions as well as for a video about how to use the GloMax®-Multi+ Detection System refer to the link in the legend of Figure 2). - Express luminescence in relative light units (RLU) (Figure 3A-B).
Note: RLU represents raw data.
- Prepare stock solutions (500x) both for luminol (LSS) and horseradish peroxidase (HPSS) in SDW. Aliquots of both LSS an HPSS can be stored at -20 °C for further uses.
- Detection of OG-triggered H2O2 production
Note: Compared to flg22 and elf18, OGs are larger negatively charged molecules. These characteristics may affect their diffusion and entry into the plant tissue generating inhomogeneous responses between treated leaf discs. To standardize the response it is necessary to vacuum infiltrate OGs into the leaf discs. Any ROS production caused by the vacuum infiltration step is assessed by the leaf discs infiltrated with water, as a control.- Prepare a luminol/peroxidase working solution (5x each); considering that you will need 50 µl per well and 1 ml of dead volume to charge the injector.
Example: For 12 wells mix 16 µl LSS with 16 µl HPSS and SDW up to 1,600 µl. - Prepare a 1.25x working solution containing OGs (OWS) in SDW; consider that you will need 200 µl per well. The final concentration of the OG solution may be comprised between 200 or 300 µg/ml.
Example: For 12 wells dissolve 0.6 mg of OGs in 1,600 µl SDW to obtain 250 µg/ml OWS. After use, discard the solution. - Gently and rapidly replace the 200 µl of water with an equal volume of OG-containing solution in each well containing leaf disc. Use 4 discs as negative control by replacing the water with the same SDW used for the OG suspension.
Note: It is not recommended to analyze more than 2 genotypes per plate, since the time required to manually replace the water with the OG solution may be too long and elicitation may occur in the firstly treated discs. In this regard it is recommended to alternate wells containing wild-type and mutant (if in analysis) discs during water replacing with the OG suspension.
Critical step: Make sure to completely submerge leaf discs in the OG solution (see step B5) prior vacuum infiltration. - Immediately vacuum infiltrate the leaf discs with the OG solution or SDW for 2 min, by using a desiccator connected to a vacuum pump.
Note: Make sure to charge the injector with the luminol/peroxidase working solution (5x each) before or during the vacuum infiltration. - Rapidly manually place the plate in the GloMax®-Multi+ Detection System. Inject 50 µl of the luminol/peroxidase working solution per well.
- Perform the luminescence detection assay for at least 40 min with a signal integration time of 1 sec.
- Express luminescence in RLU (Figure 3C).
Note: See step B7.
- Prepare a luminol/peroxidase working solution (5x each); considering that you will need 50 µl per well and 1 ml of dead volume to charge the injector.
Representative data

Figure 2. GloMax®-Multi+ Detection System with dual injectors. For a video about how to use the GloMax click on the following link: https://ita.promega.com/resources/multimedia/instruments/features-glomax-multi/. For the manufacturer’s manual click on the following link: https://www.promega.com/~/media/files/resources/protocols/technical manuals/0/glomax-multi detection system protocol.pdf.
Figure 3. ROS production in elicitors-treated Arabidopsis leaf discs. The H2O2 production was measured in Col-0 ecotype with the luminol-based assay after treatments with flg22 (100 nM) A), elf18 (100 nM) B), OGs (200 µg/ml) C). Results are average ± SE [n = 12 for elicitor treatments; n = 4 for water treatment (see Figure 3C)].
Recipes
- 500x luminol stock solution (LSS)
Dissolve 15 mg of luminol in 1 ml of DMSO, by vortexing for 10-20 sec until the solution becomes uniformly light green. After use, aliquots can be stored at -20 °C up to 3 months.
Note: Solutions are extremely sensitive to light. - 500x horseradish peroxidase stock solution (HPSS)
Dissolve 10 mg of peroxidase in 1 ml of water, by vortexing for 10-20 sec until the solution becomes uniformly brown. After use, aliquots can be stored at -20 °C up to 3 months. - 1 mM flg22 stock solution (FSS - flg22 Molecular Weight: 2272.52)
Dissolve 2.27 mg of flg22 in 1 ml of SDW, by vortexing for 10-20 sec until the solution becomes transparent.
After use, aliquots can be stored at -20 °C for 4-6 months.
Prepare the flg22 5x working solution (FWS) by diluting the FSS with SDW.
After use, discard the solution. - 1 mM elf18 stock solution (ESS - elf18 Molecular Weight: 2069.37)
Dissolve 2.07 mg of elf18 in 1 ml of SDW, by vortexing for 10-20 sec until the solution becomes transparent.
After use, aliquot and store at -20 °C for 4-6 months.
Prepare the elf18 5x working solution (EWS) by diluting the ESF with SDW.
After use, discard the solution.
References
- Bellincampi, D., Dipierro, N., Salvi, G., Cervone, F. and De Lorenzo, G. (2000). Extracellular H(2)O(2) induced by oligogalacturonides is not involved in the inhibition of the auxin-regulated rolB gene expression in tobacco leaf explants. Plant Physiol 122(4): 1379-1385.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Bisceglia, N. G., Gravino, M. and Savatin, D. V. (2015). Luminol-based Assay for Detection of Immunity Elicitor-induced Hydrogen Peroxide Production in Arabidopsis thaliana Leaves. Bio-protocol 5(24): e1685. DOI: 10.21769/BioProtoc.1685.
- Bellincampi, D., Dipierro, N., Salvi, G., Cervone, F. and De Lorenzo, G. (2000). Extracellular H(2)O(2) induced by oligogalacturonides is not involved in the inhibition of the auxin-regulated rolB gene expression in tobacco leaf explants. Plant Physiol 122(4): 1379-1385.
Category
Plant Science > Plant immunity > Disease bioassay
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










