- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
GC-MS-Based Analysis of Chloroform Extracted Suberin-Associated Root Waxes from Arabidopsis and Other Plant Species
Published: Vol 5, Iss 24, Dec 20, 2015 DOI: 10.21769/BioProtoc.1679 Views: 12407
Reviewed by: Arsalan DaudiZiqiang ZhuAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Sorghum bicolor Extracellular Vesicle Isolation, Labeling, and Correlative Light and Electron Microscopy
Deji Adekanye [...] Jeffrey L. Caplan
Oct 5, 2024 2117 Views

Analysis of Modified Plant Metabolites Using Widely Targeted Metabolite Modificomics
Jianing Zhang [...] Jun Yang
Apr 5, 2025 1469 Views

PhosphoLIMBO: An Easy and Efficient Protocol to Separate and Analyze Phospholipids by HPTLC From Plant Material
Louise Fougère [...] Yohann Boutté
Sep 5, 2025 1343 Views
Abstract
The periderm and exodermis of taproots and tuberous taproots contain an extracellular lipid polymer, suberin, deposited in their cell walls. This polymer is intractable in organic solvents, and is co-deposited with chloroform-extractable waxes. These suberin-associated root waxes are typically composed of alkanes, primary alcohols, fatty acids, alkyl ferulates, alkyl caffeates, and alkyl coumarates (Espelie et al., 1980; Li et al., 2007; Kosma et al., 2015). They are believed to contribute to the diffusion barrier properties of suberized cell walls (Soliday et al., 1979), and possibly have other roles yet to be discovered. Here we describe a protocol to extract and analyze waxes associated with root suberin. This fraction of aliphatic components is extracted by whole root immersion in chloroform, and is then chemically modified to prepare samples that are more suitable to gas-chromatography analysis. This protocol is optimized for Arabidopsis thaliana, but can be used with roots of other plants as described herein.
Materials and Reagents
- 3.5 inch pots for Arabidopsis, bigger pots if necessary for other plants
- 9 ml glass tubes with polytetrafluoroethylene (PFTE)-lined caps (Corning, catalog number: 9826-13 )
- Glass Pasteur pipets
- Glass wool (Sigma-Aldrich, catalog number: 18421 )
- 2 ml GC vials and caps (Agilent Technologies, catalog number: 5190-2240 ) with 250 µl glass inserts (Agilent Technologies, catalog number: 5181-1270 )
- 10 µl and 500 µl Hamilton syringes (Hamilton Company, catalog number: 80075 and 81216 , respectively)
Note: It is advisable to rinse the glass Hamilton syringes with acetone then chloroform before each use and chloroform then acetone after each use. This will help to prevent clogging of the syringes. - Arabidopsis thaliana or other plant seeds [e.g., Brassica napus, Raphanus sativus, Beta vulgaris, see Kosma et al., (2015) for a comprehensive list of species that have been analyzed with this protocol]
- Soil-less growing medium
- 2:1 (v/v) mixture of potting mix (Pro-Mix BX, Premier Tech Horticulture, Rivière-du-Loup)
- calcined clay granules (PPC Greens Grade, Profile, Buffalo Grove)
Note: For images of these growing media please refer to https://ag.purdue.edu/hla/Hort/greenhouse/pages/101-ways-to-grow-arabidopsis.aspx
- 2:1 (v/v) mixture of potting mix (Pro-Mix BX, Premier Tech Horticulture, Rivière-du-Loup)
- Distilled water
- Chloroform (CHCl3) (≥ 99.5% purity) (Sigma-Aldrich, catalog number: C2432 )
- 95% Ethanol (e.g., Corning, Koptec, catalog number: V1105 )
- Suggested internal standards
- Pentadecanoic acid (15:0) (Nu-Check Prep, catalog number: N-15-A ) or heptadecanoic acid (17:0) (Nu-Check Prep, catalog number: N-17-A )
- Tricosan-1-ol (23:0-OH) (Nu-Check Prep, catalog number: A-624 )
- Monoheptadecanoin (17:0 monoacylglycerol) (Nu-Check Prep, catalog number: M-159 )
- Octacosane (28:0) (Sigma-Aldrich, catalog number: O504 )
- Tridecyl (13:0) ferulate, if used need to synthesize according to Kosma et al., (2012)
- Heptadecyl (17:0) coumarate, if used need to synthesize according to Kosma et al., (2012)
- Nonadecyl (19:0) caffeate, if used need to synthesize according to Razeq et al., (2014)
- Pentadecanoic acid (15:0) (Nu-Check Prep, catalog number: N-15-A ) or heptadecanoic acid (17:0) (Nu-Check Prep, catalog number: N-17-A )
- Nitrogen gas (> 99% Purity)
- Pyridine, anhydrous (C5H5N) (Sigma-Aldrich, catalog number: 270970 )
- N, O-bis-(trimethylsilyl)-trifluoroacetamide (BSTFA) (Sigma-Aldrich, catalog number: 15222 )
- n-heptane, anhydrous (Sigma-Aldrich, catalog number: 246654 )
- Toluene, anhydrous (Sigma-Aldrich, catalog number: 244511 )
Equipment
- Controlled-environment plant growth chamber (e.g., Percival Scientific, Conviron, etc.)
- Small pair of scissors or a razor blade
- Temperature-controlled evaporator connected to nitrogen tank (e.g., Organomation Associates, catalog number: 11155 )
- Dry heating block capable of reaching a temperature of 110 ºC and with a metal block containing 13 mm orifices
- Gas chromatograph-mass spectrometer (e.g., Agilent Technologies, model: 6850/5975 GC-MS ) equipped with a HP-5MS capillary column (length 30 m, id 0.25 mm, film thickness 0.25 µm) (Agilent Technologies, J&W Scientific)
Software
- ImageJ software (National Institute of Health)
- Excel software (Microsoft office)
Procedure
- Grow plants to maturity in a controlled-environment plant growth chamber. Typical growth parameters are 21 °C to 22 °C, 40% to 60% humidity, a 16/8-h light/dark cycle, and a fluorescent light intensity of 80 to 100 μmol m-2 s-1. For Arabidopsis, seven-week-old plants should be used to allow development of mature periderm enriched in root waxes. For Arabidopsis, a density of up to four to five evenly spaced plants per pot is optimal with each replicate consisting of roots pooled from all the plants in three to four pots (12-20 plants per replicate). Four to five replicates are recommended. If multiple genotypes are employed, then a randomized complete block design should be employed to account for environmental gradients and other sources of variation. For plants with large taproots, like rapeseed (Brassica napus), each replicate can comprise one taproot from a single plant.
- The root waxes accumulate in the periderm of mature taproots (Kosma et al., 2012) and so it is convenient to cut away non-taproot tissues including secondary roots (taproots are up to 5 cm from the base of the Arabidopsis rosette; see Figure 1). For plants like rapeseed, the taproot will be longer.
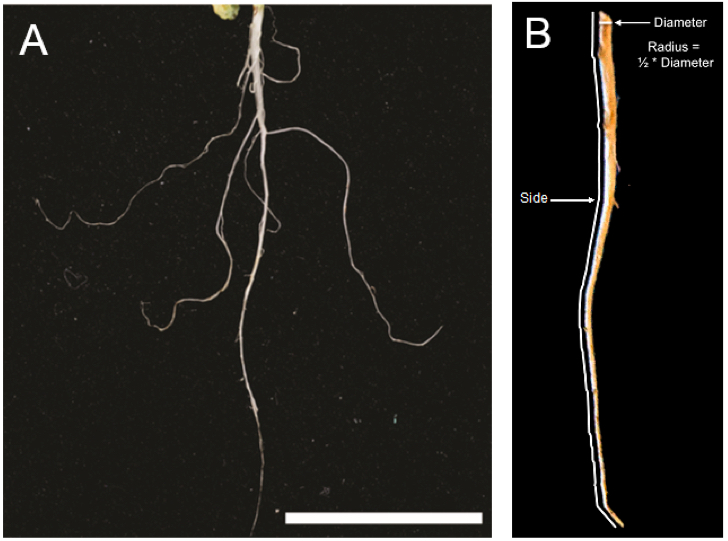
Figure 1. Arabidopsis taproot typical of what would be used for root wax extraction. A. Although lateral roots are pictured here, these should be removed prior to extraction. Scale bar = 2 cm (Kosma et al., 2015). B. Illustration of root measurements required to calculate root surface area. - Five cm of Arabidopsis taproot can be readily removed from growing media by turning the pot upside-down into your hand and removing the bulk of potting medium with a gentle stream of water or by submersion into a tub of distilled water. Lateral roots are excised with a small pair of scissors or razor blade. Root mass below 5 cm is also removed by excision with a small pair of scissors or razor blade.
- Rinse roots well with distilled water to remove remaining potting medium and debris. Gently blot dry with a paper towel. Measure and record the fresh weight.
- For Arabidopsis, place roots into a 9 ml glass tube that employs a PTFE-lined screw cap. Use a long pair of forceps to gently place roots into the glass tube. For plant species with a larger taproot like rapeseed, a glass beaker or larger glass tube should be used. All tubes or beakers must be rinsed with organic solvents to remove residual detergents and/or other sources of lipid contamination. For this, rinse tubes or beakers once with ethanol and twice with chloroform prior to use for extractions.
- Fill glass tube or beaker (containing roots) with volume of chloroform that allows complete submersion of the root(s). For Arabidopsis, between 5 and 7 ml of chloroform is sufficient (see Figure 2). Larger taproots will require a larger volume of solvent.

Figure 2. Example of a periderm wax extraction from Arabidopsis taproots. Image of a typical extraction for 1 replicate; 16 taproots submerged in 5 ml of chloroform in a 9 ml glass tube with a PTFE-lined screw cap. - Gently rotate the tube for 1.5 min with Arabidopsis or 2 min with larger taproots. If using a beaker for larger taproots, then solvent-rinsed forceps may be used to completely submerge the roots; gently agitate these roots in the beaker of chloroform.
- For Arabidopsis, decant chloroform, being careful not to remove taproots, into a solvent-rinsed 9 ml glass tube (with PTFE-lined cap). For larger taproots, the chloroform can be decanted into a large volume, solvent-rinsed tube (e.g., 25 ml). For very large volumes of chloroform (> 25 ml), solvent can be decanted into a solvent-rinsed round bottom flask (50 - 250 ml) with a ground glass neck to facilitate removal of chloroform using a rotary.
- Keep roots for surface area determination (step 17 below).
- If many particulates are visible in the extracts, extracts should be filtered through glass wool. For small volumes of extract, a small plug of glass wool can be packed into a Pasteur pipet. For larger volumes, pack glass wool into a solvent-rinsed glass funnel. Pre-rinse the glass wool with chloroform, then pour or pipet the chloroform extract through glass wool into a new, solvent-rinsed glass tube or round bottom flask.
- Add internal standards to chloroform extract using a chloroform-rinsed 10 µl Hamilton syringe. For the parameters described herein for Arabidopsis, a single, fixed concentration in the range of 5 to 10 µg of each internal standard is recommended. Larger quantities of internal standards may be required for plants like rapeseed that possess large taproots.
- For small volumes of extract (<25 ml), evaporate samples to dryness under a gentle stream of nitrogen with the bath heated to a temperature of 30 - 40 °C. For small volumes, samples will evaporate in approximately 15-20 min. For larger volumes of extract (>25 ml), round bottom flasks should be connected to a rotary evaporator under reduced pressure and with a water bath temperature of 30 - 40 °C. Once most solvent has been removed by rotary evaporation, transfer the remaining extract to a solvent-rinsed 9 ml tube and evaporate to dryness under a gentle stream of nitrogen.
- To each tube containing dry extract, add 0.5 ml of pyridine and 0.5 ml of BSTFA using a chloroform-rinsed 500 µl Hamilton syringe (Note: Volumes here can be approximate and so a Pasteur pipet can be used instead.). Cap tubes under nitrogen gas and incubate at 110 °C for 10 min using a dry heating block. This step silylates hydroxyl and carboxylate groups (derivatization) making compounds containing these groups more amenable to separation by gas chromatography.
- Evaporate derivatized samples to dryness under a gentle stream of nitrogen (no heating).
- Dissolve samples in a 1:1 mixture of n-heptane:toluene. Add 100 μl for Arabidopsis extracts. For extracts from larger taproots, greater volumes will be required.
- Transfer each sample to a GC vial containing a fresh glass insert and place on GC autosampler.
- For GC-MS analysis, the helium flow rate is 1.5 ml per minute and samples usually injected in splitless mode. For typical root wax samples, the temperature settings are: inlet at 350 °C; detector at 320 °C; oven program at 130 °C for 2 min, increased to 325 °C at 5 °C per min, and then held at 325 °C for 10 min.
- To calculate root surface area (SA), each whole root is scanned along its longitudinal axis using a desktop scanner. Image files should be scanned at a resolution of 200 dpi (78.74 dots per cm) and saved as TIFF files. The digital TIFF files are opened in ImageJ and the built-in software tools used to determine radius (r) of the cut base and length of the side of the root (s) (Figure 1B). s is taken as the mean length of two sides from the digitized two-dimensional image and r calculated as ½ the measured width of the cut base. The surface areas of the taproots are then calculated using the formula SA = π * r * s (a conical geometry is assumed).
- A typical chromatogram of an Arabidopsis taproot wax extract is shown in Figure 3. The amount (μg) of each constituent is calculated from its peak area relative to the peak area of the respective internal standard. Fatty alcohol peaks should be compared to the tricosan-1-ol internal standard, alkyl coumarate peaks to the heptadecyl coumarate internal standard, etc. Constituent amounts (χ μg) are calculated by multiplying the internal standard (IS) amount (μg) with the peak area (PA) of the constituent of interest (COI) and dividing by the peak area of the internal standard. In the following formula, 10 μg of IS is used: χ μg = 10 μg * PACOI/PAIS. The following is an example using 10 micrograms of IS, PACOI of 25,000 (arbitrary units), and a PAIS of 12,000: χ μg = 10 μg * 25,000 area units/12,000 area units, χ μg = 10 μg * 2.0833, χ = 20.833 μg. The wax constituent amounts are then divided by total surface area or fresh weight (FW) of root(s) used for extraction (e.g., µg cm-2 or µg g-1 FW). Continuing with the above example, if one replicate consists of 100 mg of FW and our peak amount is 20.833 μg then 20.833 μg/100 mg FW = 0.20833 μg mg-1 FW. However, surface area is generally preferred because the root waxes are being extracted from surface tissue layer (periderm), thereby allowing for better comparisons between species of differing root size (surface area to fresh weight ratios vary dramatically).
Representative data
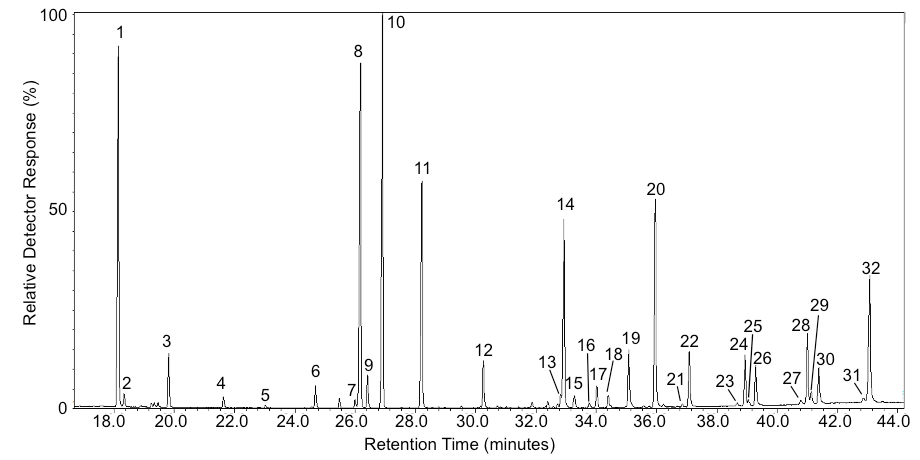
Figure 3. Example chromatogram of Arabidopsis root waxes. Peak IDs are as follows: 1) 17:0 FFA*; 2) 18:0-OH; 3) 18:0 FFA; 4) 20:0-OH; 5) 20:0 FFA; 6) 22:0-OH; 7) 22:0 FFA; 8) 23:0-OH*; 9) β-17:0 MAG*; 10) α-17:0 MAG*; 11) 28:0 Alkane*; 12) cis-13:0 ferulate*; 13) β-20:0 MAG; 14) trans-13:0 ferulate*; 15) α-20:MAG; 16) cis-17:0 coumarate*; 17) 28:1 sterol; 18) 29:2 sterol; 19) 29:1 sterol; 20) trans-17:0 coumarate*; 21) cis-18:0 caffeate; 22) trans-18:0 coumarate; 23) trans-18:0 ferulate; 24) trans-18:0 caffeate; 25) cis-20:0 caffeate; 26) trans-20:0 coumarate; 27) trans-20:0; ferulate; 28) trans-20:0 caffeate; 29) cis-22:0 caffeate; 30) trans-22:0 coumarate; 31) trans-22:0 ferulate; 32) trans-22:0 caffeate. Abbreviations are as follows: FFA = free fatty acid, -OH = primary fatty alcohol, MAG = monoacylglycerol, * indicates an internal standard (used for quantification).
Notes
- The soil-less medium used here leaves little debris on the roots and thus the roots are easily cleaned prior to chloroform extraction. Other growing media, including soil, can be used instead but then it may be more difficult to wash material away from the roots.
- It is important that no plasticware, such as plastic pipettemen tips, are used during the extraction and processing of root waxes or else there is a good chance of plastic contamination in the GC traces. Use solvent rinsed glassware and Hamilton syringes throughout the procedure. New glass Pasteur pipets do not need to be rinsed with solvent.
- The optimum mix of internal standards depends on the chemical composition of root waxes. The mix recommended herein is ideal for Arabidopsis root waxes. For example, not all species contain alkyl caffeates. As such, the nonadecyl caffeate internal standard could then be excluded. It is important that the internal standards used do not co-elute with any of the native root wax components. Each root wax component should be quantified relative to an internal standard of like chemical structure (i.e., they only differ in chain length).
- Root waxes are influenced by stage of development (e.g., may accumulate only in mature periderm) and by environment (e.g., may be stress induced). Therefore, one needs to ensure that control and sample plants (e.g., a mutant) are grown together and sampled exactly the same way.
- Although the protocol described here is optimized for Arabidopsis root waxes, it can be modified for any other plant by taking into account differing root sizes and root wax compositions, as noted above and, for example, as reported in Razeq et al. (2014) and Kosma et al. (2015).
Acknowledgments
The original version of this protocol was reported in Li et al. (2007) with adjustments made in subsequent publications (Molina et al., 2009; Kosma et al., 2012; Vishwanath et al., 2013) to produce the protocol reported here.
References
- Espelie, K. E., Sadek, N. Z. and Kolattukudy, P. E. (1980). Composition of suberin-associated waxes from the subterranean storage organs of seven plants: Parsnip, carrot, rutabaga, turnip, red beet, sweet potato and potato. Planta 148(5): 468-476.
- Kosma, D. K., Molina, I., Ohlrogge, J. B. and Pollard, M. (2012). Identification of an Arabidopsis fatty alcohol:caffeoyl-Coenzyme A acyltransferase required for the synthesis of alkyl hydroxycinnamates in root waxes. Plant Physiol 160(1): 237-248.
- Kosma, D. K, Rice, A. and Pollard M. (2015). Analysis of aliphatic waxes associated with root periderm or exodermis from eleven plant species. Phytochemistry 117: 351-362.
- Li, Y., Beisson, F., Ohlrogge, J. and Pollard, M. (2007). Monoacylglycerols are components of root waxes and can be produced in the aerial cuticle by ectopic expression of a suberin-associated acyltransferase. Plant Physiol 144(3): 1267-1277.
- Molina, I., Li-Beisson, Y., Beisson, F., Ohlrogge, J. B. and Pollard, M. (2009). Identification of an Arabidopsis feruloyl-coenzyme A transferase required for suberin synthesis. Plant Physiol 151(3): 1317-1328.
- Razeq, F. M., Kosma, D. K., Rowland, O. and Molina, I. (2014). Extracellular lipids of Camelina sativa: characterization of chloroform-extractable waxes from aerial and subterranean surfaces. Phytochemistry 106: 188-196.
- Soliday, C. L., Kolattukudy, P. E., and Davis, R. W. (1979). Chemical and ultrastructural evidence that waxes associated with the suberin polymer constitute the major diffusion barrier to water vapor in potato tuber (Solanum tuberosum L.). Planta 146(5): 607-614.
- Vishwanath, S. J., Kosma, D. K., Pulsifer, I. P., Scandola, S., Pascal, S., Joubes, J., Dittrich-Domergue, F., Lessire, R., Rowland, O. and Domergue, F. (2013). Suberin-associated fatty alcohols in Arabidopsis: distributions in roots and contributions to seed coat barrier properties. Plant Physiol 163(3): 1118-1132.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Kosma, D. K., Molina, I. and Rowland, O. (2015). GC-MS-Based Analysis of Chloroform Extracted Suberin-Associated Root Waxes from Arabidopsis and Other Plant Species. Bio-protocol 5(24): e1679. DOI: 10.21769/BioProtoc.1679.
Category
Plant Science > Plant biochemistry > Lipid
Plant Science > Plant metabolism > Metabolite profiling
Plant Science > Plant physiology > Tissue analysis
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link












