- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Purification of Bacterial RNA from Infected Macrophages
Published: Vol 5, Iss 22, Nov 20, 2015 DOI: 10.21769/BioProtoc.1660 Views: 11195
Reviewed by: Fanglian HeYoko Eguchi

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Quantification of Colibactin-associated Genotoxicity in HeLa Cells by In Cell Western (ICW) Using γ-H2AX as a Marker
Sophie Tronnet and Eric Oswald
Mar 20, 2018 8082 Views

Adhesion of Enteroaggregative E. coli Strains to HEK293 Cells
Jorge Luis Ayala-Lujan and Fernando Ruiz-Perez
Apr 20, 2018 6852 Views

Intracellular Invasion and Killing Assay to Investigate the Effects of Binge Alcohol Toxicity in Murine Alveolar Macrophages
Victor Jimenez Jr and Fernando P Monroy
Jan 20, 2019 6741 Views
Abstract
Studying the transcriptome of bacterial pathogens during infection is a very informative and effective tool for discovering genes that contribute to successful infection. However, isolating bacterial RNA from infected cells or tissues is a challenging process due to the much higher amounts of host RNA in the lysates of infected cells. We have optimized a method for isolating RNA of Listeria monocytogenes (L. monocytogenes) bacteria infecting bone marrow derived macrophage cells (BMDM). After infection, we lyse the cells and filter the lysates through 0.45 µm filters to discard most of the host proteins and RNA. Next, we resuspend the bacteria and extract RNA following DNase treatment. The extracted RNA is suitable for gene expression analysis by real-time PCR or microarray. We have successfully employed this protocol in our studies of Listeria monocytogenes gene regulation during infection in vitro (Lobel et al., 2015; Lobel et al., 2012; Kaplan Zeevi et al., 2013; Rabinovich et al., 2012).
Keywords: Listeria monocytogenesMaterials and Reagents
- Cell scrapers (Thermo Fisher Scientific, NuncTM, catalog number: 179693 )
- MF-Millipore filters (Merck Millipore Corporation, catalog number: HAWP04700 )
- Cell culture dishes (Greiner Bio One International GmbH, catalog number: 639160 )
- Eppendorf tubes (Corning Inc., Axygen, catalog number: MCT175C )
- Pipettes sterile (25 ml) (Greiner Bio One International GmbH)
- Falcon tubes (50 ml) (Greiner Bio One International GmbH)
- Listeria monocytogenes 10403S (Daniel Portnoy’s lab stock) (Becavin et al., 2014)
- Bone marrow (from C57B/6 female mice; ordered from Harlan labs Israel)
- Liquid nitrogen
- RNase-free water (Thermo Fisher Scientific, InvitrogenTM, catalog number: 10977015 )
- DMEM (Thermo Fisher Scientific, GibcoTM, catalog number: 41965039 )
- L-Glutamine (200 mM) (Thermo Fisher Scientific, GibcoTM, catalog number: 25030081 )
- Sodium pyruvate (Thermo Fisher Scientific, GibcoTM, catalog number: 11360088 )
- 2-Mercaptoethanol (Thermo Fisher Scientific, GibcoTM, catalog number: 31350010 )
- Penicillin-Streptomycin (10,000 U/ml) (Thermo Fisher Scientific, GibcoTM, catalog number: 15140122 )
- Gentamicin (Sigma-Aldrich, catalog number: G1397 )
- Fetal Bovine Serum (FBS) (Thermo Fisher Scientific, GibcoTM, catalog number: 10270106 )
- Dulbecco’s Phosphate Buffered Saline (Sigma-Aldrich, catalog number: D8537 )
- Brain Heart Infusion (BHI) (Merck Millipore Corporation, catalog number: 1104930500 )
- Phenol saturated (pH 4.3) (Thermo Fisher Scientific, catalog number: BP1751I-400 )
- Chloroform (Thermo Fisher Scientific, catalog number: BP1145-1 )
- Isoamyl alcohol (Sigma-Aldrich, catalog number: W205702 )
- Sodium acetate Anhydrous (Sigma-Aldrich, catalog number: W302406 )
- Ethylenediaminetetraacetic acid (EDTA) (Sigma-Aldrich, catalog number: EDS )
- DNase I, RNase-free (supplied with MnCl2) (1 U/µl) (Fermentas, catalog number: EN0521 )
Note: Currently, it is “Thermo Fisher Scientific, catalog number: EN0521”. - 10% Sodium dodecyl sulfate (SDS) (Sigma-Aldrich, catalog number: L4522 )
- Ethanol absolute (Merck Millipore Corporation, catalog number: 1070174000 )
- M-CSF (L-929 conditioned medium) (Englen et al., 1995)
Note: Alternatively M-CSF can be bought from Sigma (Sigma-Aldrich, catalog number: SRP3221 ). - BMDM + PS media (filter sterilized) (see Recipes)
- BMDM no PS media (filter sterilized) (see Recipes)
- AE buffer (see Recipes)
- Phenol-chloroform-IAA (see Recipes)
- Chloroform-IAA (see Recipes)
Equipment
- CO2 forced-air incubator (Thermo Fisher Scientific, model: 3111 )
- Kontes glass holder (Thermo Fisher Scientific, catalog number: K953755-0045 )
- Speed-Vac Concentrator (Thermo Fisher Scientific, catalog number: SPD131DDA )
- Vortex-Genie 2 (Scientific Industries, model: G560E )
- Nanodrop 1000 (Thermo Fisher Scientific)
- 30 °C incubator (Thermo Fisher Scientific)
- 65 °C heat block (Thermo Fisher Scientific)
- 4 °C table centrifuge (Thermo Fisher Scientific, EppendorfTM, model: 5417R )
Procedure
- Infection
Day 1:- Seed 2.0 x 107 bone marrow derived macrophages (BMDM) in 145 mm dishes with 30 ml BMDM + PS media, 3 plates for each bacterial strain being analyzed.
- Pick a bacterial colony from a BHI plate and inoculate into 10 ml of BHI medium (repeat for each sample), incubate at 30 °C incubator, slanted without shaking overnight.
- Pre-warm BMDM no PS medium and PBS at 37 °C.
- Wash macrophage monolayer twice with 25 ml of warmed PBS to remove the antibiotics.
- Add 30 ml fresh medium.
- Wash 1.5 ml of overnight bacterial cultures twice with PBS.
- Infect macrophages at MOI of 90:1 (in favor for the bacteria) for wild-type L. monocytogenes 10403S. When mutants defected in intracellular growth are used, consider increasing the MOI.
- Infect each plate 15 min apart (same as in step A5).
- Following 0.5 h incubation (in respect to each sample infection time), wash monolayers twice with 30 ml PBS. Add 30 ml pre-warmed BMDM no PS medium.
- At 1 h post infection, add 30 µl of gentamicin (1:1,000) to kill extracellular bacteria.
- Prepare filter apparatus and ice-cold RNase-free water.
- At 6 h post infection, harvest the bacteria. Treat each plate individually.
- Wash monolayers with PBS once.
- Add 20 ml of ice-cold RNase-free water.
- Using a cell scraper, scrape and lyse cells by pushing the liquid in front of the scraper.
- Collect by pipetting the lysed monolayer in a 50ml conical tube. Vortex for 30 sec. Centrifuge 2,000 rpm (720 x g) /3 min/ 4 °C.
- Pass supernatant through filter apparatus. Using tweezers, put the filter membrane in 15 ml conical tubes, loosely rolled.
- Immediately freeze filters by dipping the 15 ml tubes in liquid nitrogen.
- Prepare the filter apparatus for the next sample and repeat.
- Wash monolayers with PBS once.
- Seed 2.0 x 107 bone marrow derived macrophages (BMDM) in 145 mm dishes with 30 ml BMDM + PS media, 3 plates for each bacterial strain being analyzed.
- Nucleic acids extraction
Day 3:- Prepare 1:1 mix of acidified phenol:chloroform, 400 µl for each sample. Mix in a separate tube, and then expel the resulting aqueous layer aspirated off.
- Thaw your filter-containing tubes on ice. Insert each tube fully into ice.
- To each filter-containing tube add 650 µl AE buffer. Add buffer to all tubes.
- Working quickly, vigorously vortex your filter containing tube so that the filter whisks to the periphery of the tube and the buffer fully washes over the filter. It may be necessary to additionally vortex the tube while inverted to fully wash the bacteria off the filter. Always keep tube cold by placing it back on ice. Repeat for all tubes. It may be useful to spin-down the suspension shortly (1 min/750 rpm/60 x g) at the end of the process.
- Transfer the bacteria-containing buffer to the 1.5 ml Eppendorf tube containing 40 µl of 10% SDS and 400 µl of phenol/chloroform mix. Repeat for all tubes. It may be necessary to spin down tubes shortly (1 min/750 rpm) to get residual buffer from the filter.
- Place all of your tubes in the multi-tube vortex device, and vortex at full speed for 10 min.
- Incubate the tubes at 65 °C heat block for 10 min.
- Centrifuge at maximum speed (14,000 RPM / 20,817 x g) for 5 min.
- Transfer the aqueous layer (about 400 µl) from each tube to the 1.5 ml tube containing 40 µl 3 M sodium acetate (pH 5.2) and 1.0 ml 100% ethanol. Vortex each tube thoroughly.
- Incubate your sample at -80 °C 1 h.
- Centrifuge at 4 °C for 20 min at maximum speed (14,000 RPM / 20,817 x g).
- Carefully aspirate off the ethanol from each tube. Add 500 µl cold 70% ethanol to each sample and vortex thoroughly.
- Centrifuge at 4 °C for 20 min at maximum speed.
- Carefully aspirate off the ethanol from each tube. Dry your samples for approximately 2 min in a speed-Vac device without heating. It is critical not to over-dry your samples.
- To each sample, add 25 µl RNase-free water. Incubate at room temperature for 20 min. Carefully vortex and spin-down.
- Analyze the RNA concentration in the samples using the NanoDrop. Expect to extract about 0.5-1 µg/plate.
- You can unite the technical replicates at this stage.
- Prepare 1:1 mix of acidified phenol:chloroform, 400 µl for each sample. Mix in a separate tube, and then expel the resulting aqueous layer aspirated off.
- DNase treatment
- Set reaction:
RNA up to 2 µg (max 44 µl) 10x DNase buffer 5 µl H2O complete to 50 µl DNase 1 µl Total 50 µl - Incubate at 37 °C for 45 min.
- Add 450 µl RNase-free water and 500 µl of Phenol-Chloroform-IAA mix, separate phases by 2 min centrifugation at maximal speed.
- Transfer the aqueous layer to a new tube and add 500 µl chloroform-IAA mix, vortex and separate phases by 2 min centrifugation at maximal speed.
- Transfer the aqueous layer to a new tube and add 1 ml of ethanol and 50 µl of 3 M sodium acetate (pH 5.2). Vortex.
- Incubate for 1 h at -80 °C.
- Centrifuge at 4 °C for 20 min at maximal speed.
- Carefully aspirate off the ethanol from each tube. Add 500 µl cold 75% ethanol to each sample and vortex thoroughly.
- Centrifuge at 4 °C for 20 min at maximum speed.
- Carefully aspirate off the ethanol from each tube. In a speed-Vac without heating, dry your samples for approximately 2 min. It is critical not to over-dry your samples.
- Add 12 µl of RNase-free water, incubate for 2 min at room temperature, vortex, and spin-down. Keep the RNA on ice.
- Analyze the RNA concentration in the samples using the NanoDrop. Expect to extract about 100 ng/sample.
- Set reaction:
Representative data
We provide representative data of induction of the virulence gene hly of L. monocytogenes during intracellular growth in macrophages compared to growth in BHI medium (Figure 1). mRNA was purified from intracellular bacteria 6 hours post infection at M.O.I of 90:1 and from bacteria grown to mid log (OD600 = 0.5) in BHI medium. 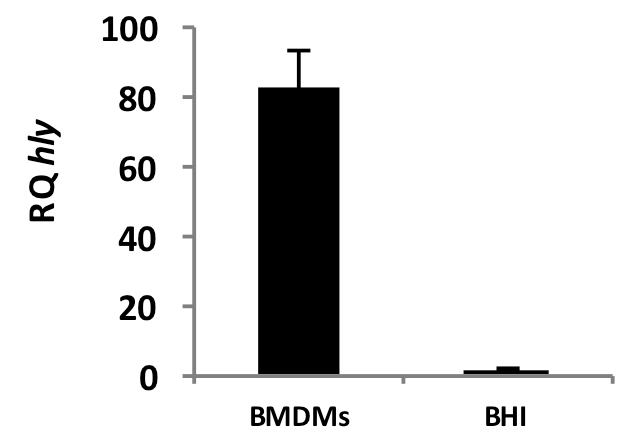
Figure 1. Transcription of hly during growth of L. monocytogenes in BMDMs (6 h.p.i.) and BHI lab medium. Error bars represent 95% confidence interval. Results are representative of 3 repeats (N=3). RQ: Relative quantity
Notes
- Check the morphology of the infected cells before and after infection. The cells should be viable during the infection process.
- Be very careful when handling the membranes-use tweezers and touch only the edges.
- When passing the samples through the filter apparatus, first filter RNase-free water to verify that the vacuum is working and that there is no blockage in the system.
- You should observe a reduction in nucleic acid concentration following DNase I treatment. If you don’t observe a reduction, consider another round of DNase I treatment.
Recipes
- BMDM+PS media (filter sterilized)
DMEM 250 ml FBS (inactivated 30 min at 54 °C) 100 ml M-CSF (L-929 conditioned medium) (Englen et al., 1995) 150 ml Glutamine 5 ml Sodium pyruvate 5 ml β-Mercaptoethanol 0.5 ml Pen/Strep 5 ml - BMDM no PS media (filter sterilized)
DMEM 255 ml FBS (inactivated 30 min at 54 °C) 100 ml M-CSF (L-929 conditioned medium) (Englen et al., 1995) 150 ml Glutamine 5 ml Sodium pyruvate 5 ml β-Mercaptoethanol 0.5 ml - AE buffer
50 mM NaOAc (pH 5.2)
10 mM EDTA
RNase-free water - Phenol-chloroform-IAA
Phenol 25 ml
Chloroform 24 ml
Isoamyl alcohol 1 ml - Chloroform-IAA
Chloroform 24 ml
Isoamyl alcohol 1 ml
Acknowledgments
This protocol was adapted from the previously published studies (Lobel et al., 2015; Kaplan Zeevi et al., 2013; Lobel et al., 2012). This work was supported by the Israel Science Foundation and the ERA-NET PathoGenoMics (funded by MOH Israel) grants to AAH. L.L is supported by the Argentinian friends of TAU.
References
- Becavin, C., Bouchier, C., Lechat, P., Archambaud, C., Creno, S., Gouin, E., Wu, Z., Kuhbacher, A., Brisse, S., Pucciarelli, M. G., Garcia-del Portillo, F., Hain, T., Portnoy, D. A., Chakraborty, T., Lecuit, M., Pizarro-Cerda, J., Moszer, I., Bierne, H. and Cossart, P. (2014). Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. MBio 5(2): e00969-00914.
- Englen, M. D., Valdez, Y. E., Lehnert, N. M. and Lehnert, B. E. (1995). Granulocyte/macrophage colony-stimulating factor is expressed and secreted in cultures of murine L929 cells. J Immunol Methods 184(2): 281-283.
- Kaplan Zeevi, M., Shafir, N. S., Shaham, S., Friedman, S., Sigal, N., Nir Paz, R., Boneca, I. G. and Herskovits, A. A. (2013). Listeria monocytogenes multidrug resistance transporters and cyclic di-AMP, which contribute to type I interferon induction, play a role in cell wall stress. J Bacteriol 195(23): 5250-5261.
- Lobel, L., Sigal, N., Borovok, I., Belitsky, B. R., Sonenshein, A. L. and Herskovits, A. A. (2015). The metabolic regulator CodY links Listeria monocytogenes metabolism to virulence by directly activating the virulence regulatory gene prfA. Mol Microbiol 95(4): 624-644.
- Lobel, L., Sigal, N., Borovok, I., Ruppin, E. and Herskovits, A. A. (2012). Integrative genomic analysis identifies isoleucine and CodY as regulators of Listeria monocytogenes virulence. PLoS Genet 8(9): e1002887.
- Rabinovich, L., Sigal, N., Borovok, I., Nir-Paz, R. and Herskovits, A. A. (2012). Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 150(4): 792-802.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Lobel, L., Sigal, N., Pasechnek, A. and Herskovits, A. A. (2015). Purification of Bacterial RNA from Infected Macrophages. Bio-protocol 5(22): e1660. DOI: 10.21769/BioProtoc.1660.
Category
Microbiology > Microbial genetics > RNA > RNA extraction
Microbiology > Microbe-host interactions > In vitro model > Cell line
Molecular Biology > RNA > RNA extraction
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









