- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Expression, Purification and in vitro Enzyme Activity Assay of Plant Derived GTPase
Published: Vol 5, Iss 22, Nov 20, 2015 DOI: 10.21769/BioProtoc.1651 Views: 10784
Reviewed by: Ru ZhangAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Immunofluorescence for Detection of TOR Kinase Activity In Situ in Photosynthetic Organisms
Ana P. Lando [...] Giselle M. A. Martínez-Noël
Dec 20, 2024 1825 Views

An Activity-Based Proteomics with Two-Dimensional Polyacrylamide Gel Electrophoresis (2D-PAGE) for Identifying Target Proteases in Arabidopsis Apoplastic Fluid
Sayaka Matsui and Yoshikatsu Matsubayashi
Mar 5, 2025 1990 Views

Advancing 2-DE Techniques: High-Efficiency Protein Extraction From Lupine Roots
Sebastian Burchardt [...] Emilia Wilmowicz
Oct 5, 2025 1772 Views
Abstract
Based on gene expression data after biotic stress, the GTPase RabA4c has been suggested to regulate pathogen-induced callose biosynthesis in the model organism Arabidopsis thaliana. We studied the function of RabA4c in its native and dominant negative (dn) isoform. In planta, RabA4c overexpression prevented penetration of the virulent powdery mildew Golovinomyces cichoracearum into epidermal leaf cells. This penetration resistance was caused by enhanced callose deposition at sites of attempted fungal penetration at early time points of infection. By contrast, RabA4c (dn) overexpression did not increase callose deposition or penetration resistance.
In this protocol, we describe the expression, purification and activity assay of the heterologously expressed GTPase RabA4c from A. thaliana based on the publication Ellinger et al. (2014). We fused RabA4c to the fluorophore mCitrine and expressed this protein in the yeast strain Pichia pastoris GS115. For purification of RabA4c, we used the GFP-Trap_A kit (Chromo Tek) which specifically binds to GFP derivatives like mCitrine. The enzyme activity assay was done by using the GTPase Assay Kit from Innova Biosciences. In general, we followed the instructions made by the manufacturers.
Materials and Reagents
- Protein expression
- pGAPZ A, B, & C Pichia pastoris Expression Vectors (Life Technologies, catalog number: V200-20 )
Note: Currently, it is “Thermo Fisher Scientific, InvitrogenTM, catalog number: V200-20”. - ZeocinTM (InvivoGen, catalog number: ant-zn-5b ) for selection
- 1 % yeast extract (Carl Roth GmbH + Co., catalog number: 2363.3 )
- 2 % peptone (BD, catalog number: 211677 )
- D(+)-Glucose (Carl Roth GmbH + Co., catalog number: X997.3 )
- Yeast Peptone Dextrose (YPD) media (see Recipes)
- pGAPZ A, B, & C Pichia pastoris Expression Vectors (Life Technologies, catalog number: V200-20 )
- Protein purification
- Acid washed glass beads (Sigma-Aldrich, catalog number: G 8772 )
- GFP-Trap® (ChromoTek GmbH, catalog number: gta-20 )
- NanoOrange Protein Quantification Kit (Invitrogen, catalog number: N-6666 )
Note: Currently, it is “Thermo Fisher Scientific, Molecular ProbesTM, catalog number: N-6666”. - 10 mM Tris (pH 7.5)
- 50 mM NaCl
- 0.5 mM EDTA (pH 8.0)
- 100 mM Glycine (pH 2.5)
- 1 M Tris (pH 10.4)
- Dilution buffer (see Recipes)
- Wash buffer I (see Recipes)
- Wash buffer II (see Recipes)
- Elution buffer (see Recipes)
- Acid washed glass beads (Sigma-Aldrich, catalog number: G 8772 )
- GTPase activity assay
- GTPase Assay Kit (Innova Biosciences Ltd., catalog number: 602-0121 )
- Negative control: expressed and purified GFP using P. pastoris as expression system (plasmid pGAPZ::eGFP)
- GTPase Assay Kit (Innova Biosciences Ltd., catalog number: 602-0121 )
Equipment
- 300 ml flasks
- Shaker (160 rpm)
- Centrifuge for 50 ml Falcon tubes (3,220 x g)
- Vortex
- Tube rotator at 4 °C and room temperature
- Plate Reader (BioTek Instruments, model: Synergy HTX Multi-Mode Reader)
- Tabletop centrifuge (cooling)
Procedure
- Protein expression
- Strain: Pichia pastoris GS115
- Expression vector: pGAPZ
- Pre-culture: incubate one colony from a YPD (+ zeocin) plate in 4 ml liquid YPD (+ zeocin) media at 28 °C and 160 rpm for one day.
- Main-culture: Incubated 1 ml of the pre-culture in 100 ml fresh YPD (+ zeocin) media at 28 °C and 160 rpm for 3 days.
- Strains: pGAPZ::RabA4c-mCit[NA]/ [DN], control pGAPZ:e:GFP [NA= native form, DN= dominant negative, please refer to Ellinger et al. (2014) for strain generation].
- Yeast cells were transferred in 50 ml Falcon tubes and harvested by centrifugation (3,220 x g, 10 min, 4 °C).
- Discard the supernatant and wash the cell pellet with sterile ddH2O.
- Mechanical lysis: Add an identical volume of sterile acid washed glass beads to the cell pellet and vortex the beads-cell suspension for 30 sec at maximum speed followed by chilling on ice for 30 sec, repeat these steps A8-10 times.
- Spin down the glass beads at 453 x g at 4 °C for 2 min.
- Transfer the supernatant in a new 2 ml tube, wash the glass beads with 1 ml dilution buffer (see above), centrifuge again and transfer the supernatant also to the 2 ml tube.
- Separate the cell debris by centrifugation at max speed at 4 °C for 30 min.
- Transfer the clear supernatant in a new tube and store at 4 °C until starting with protein purification. Store the pellet at -20 °C for SDS-PAGE analysis.
- Strain: Pichia pastoris GS115
- Protein purification
- Equilibrate 25 µl (for one sample) GFP-Trap_A agarose beads in dilution buffer (use a 2 ml tube): Wash 3 times with 500 µl ice cold dilution buffer, between the wash steps spin beads down at 719 x g at 4 °C for 2 min and discard the supernatant.
- Add the crude extract to the equilibrate beads and incubate the sample for 4 h at 4 °C under constant mixing (e.g., tube rotator).
- Spin down sample at 719 x g at 4 °C for 2 min and carefully remove the supernatant by pipetting. Store an aliquot for “non-bound proteins” analysis at 4 °C.
- Wash beads 3 times with 1 ml ice cold wash buffer I and II through centrifugation at 719 x g at 4 °C for 2 min; discard the supernatant.
- Beads fused to the protein of interest are ready-to-use for enzyme activity assay.
- Equilibrate 25 µl (for one sample) GFP-Trap_A agarose beads in dilution buffer (use a 2 ml tube): Wash 3 times with 500 µl ice cold dilution buffer, between the wash steps spin beads down at 719 x g at 4 °C for 2 min and discard the supernatant.
- GTPase activity assay (for more information, please refer to the GTPase Assay Kit Manual)
GTPases [also known as G proteins (guanine nucleotide-binding proteins)] function as molecular switches that cycle between an active and inactive state. The cycle is linked to the binding and hydrolysis of GTP. In the active state, these enzymes interact with GTP to perform diverse cellular functions. Inactivation occurs through the association of GAP (GTPase-activating protein). This protein hydrolyzes GTP to GDP and free phosphate. To return to the active state, the enzyme GEF (guanine nucleotide exchange factor) is required to catalyze the exchange of GDP for GTP (Figure 1).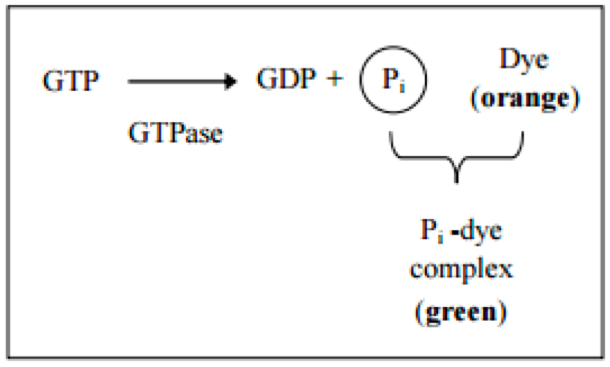
Figure 1. Principle of the GTPase assay kit (Innova Bioscience)- Prepare the substrate buffer mix (SB-Mix) and samples for a standard curve as described in the GTPase Assay kit Manual.
- Add 350 µl SB-Mix buffer to the purified beads-protein solution.
- Incubate samples at room temperature for 30 min under constant mixing.
- Spin down beads at 1,073 x g for 5 min using a tabletop centrifuge.
- Put 100 µl of reaction buffer in one well of a 96-well plate (triplicates) and add 100 µl pure H2O; prepare samples for the standard curve in the same way.
- Prepare the Gold-Mix buffer (see GTPase Assay kit Manual) and add 50 µl to each well to stop the reaction.
- After 2 min add 20 µl stabilizer.
- Incubate plate at room temperature for 30 min in the dark.
- Read plate at a wavelength of 650 nm (590- 660 nm, see GTPase Assay kit Manual).
- For calculation of enzyme activity, follow Appendix 1 of the GTPase Assay kit Manual (Figure 2).
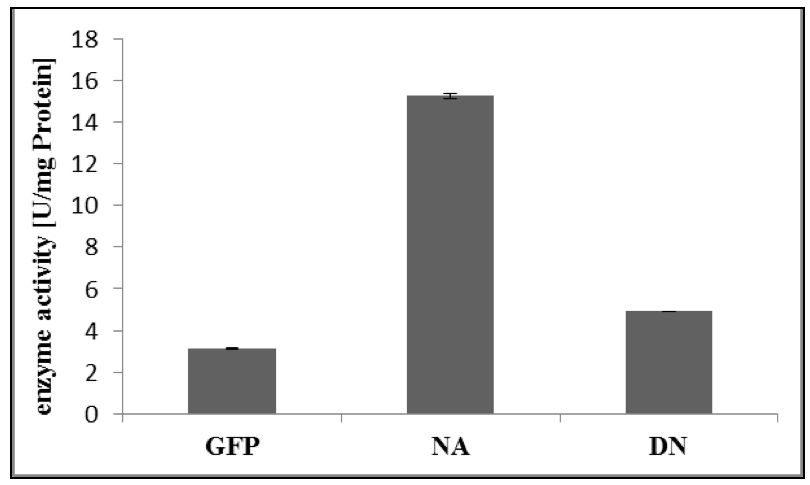
Figure 2. Specific enzyme activity of RabA4c-mCit [NA]/ [DN] and GFP as control. The GTPase assay was performed as described above (Procedure). Error bars represent standard deviations.
- Prepare the substrate buffer mix (SB-Mix) and samples for a standard curve as described in the GTPase Assay kit Manual.
- SDS-PAGE and protein quantification
- After GTPase Assay: Elute the target protein from the GFP-Trap_A beads by adding 200 µl 100 mM glycine (pH 2.5), mix shortly and incubate at room temperature for 2 min.
- Add 20 µl 1 M TRIS base (pH 10.4) for neutralization.
- Spin beads down at 1,073 x g at room temperature for 2 min, transfer supernatant to a new tube.
- Protein quantification follows the instructions of the manufactures (NanoOrange Protein Quantification Kit, see above), the protein amount is necessary to determine the enzyme activity.
- After GTPase Assay: Elute the target protein from the GFP-Trap_A beads by adding 200 µl 100 mM glycine (pH 2.5), mix shortly and incubate at room temperature for 2 min.
Recipes
- Yeast Peptone Dextrose (YPD) media
1% yeast extract
2% peptone
D(+)-Glucose - Dilution buffer
10 mM Tris (pH 7.5)
150 mM NaCl
0.5 mM EDTA (pH 8.0) - Wash buffer I
10 mM Tris (pH 7.5)
300 mM NaCl
0.5 mM EDTA (pH 8.0) - Wash buffer II
10 mM Tris (pH 7.5)
500 mM NaCl
0.5 mM EDTA (pH 8.0) - Elution buffer
100 mM glycine (pH 2.5)
1 M Tris (pH 10.4)
Acknowledgments
This work was performed as a part of the publication of Ellinger et al. (2014) and was funded by the Federal Ministry of Education and Research (BMBF) within the research focus BioEnergy 2021: "CallBio - Resistant Plants for simplified Bio-ethanol Production through optimized Biosynthesis of the Cell Wall Polymer Callose".
References
- Ellinger, D., Glöckner, A., Koch, J., Naumann, M., Stürtz, V., Schütt, K., Manisseri, C., Somerville, S. C. and Voigt, C. A. (2014). Interaction of the Arabidopsis GTPase RabA4c with its effector PMR4 results in complete penetration resistance to powdery mildew. Plant Cell 26(7): 3185-3200.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Glöckner, A. and Voigt, C. A. (2015). Expression, Purification and in vitro Enzyme Activity Assay of Plant Derived GTPase. Bio-protocol 5(22): e1651. DOI: 10.21769/BioProtoc.1651.
Category
Plant Science > Plant biochemistry > Protein > Activity
Plant Science > Plant biochemistry > Protein > Isolation and purification
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link