- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Determination of Mannitol-2-dehydrogenase Activity
Published: Vol 5, Iss 21, Nov 5, 2015 DOI: 10.21769/BioProtoc.1634 Views: 9458
Reviewed by: Valentine V TrotterChristian RothAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Surface Plasmon Resonance for the Interaction of Capsular Polysaccharide (CPS) With KpACE
Zhe Wang [...] Chao Cai
Jun 20, 2025 3554 Views

Activation of X-Succinate Synthases for Fumarate Hydroalkylation Using an In Vitro Activation Method
Anshika Vats [...] Mary C. Andorfer
Jun 20, 2025 2519 Views

An Optimized Enzyme-Coupled Spectrophotometric Method for Measuring Pyruvate Kinase Kinetics
Saurabh Upadhyay
Aug 20, 2025 2454 Views
Abstract
Mannitol is a polyol that occurs in a wide range of living organisms, where it fulfills different physiological roles. Several pathways have been described for the metabolism of mannitol by bacteria, including the phosphoenolpyruvate-dependent phosphotransferase system (PST) and a M2DH-based catabolic pathway. The latter involves two enzymes, a mannitol-2-dehydrogenase (EC 1.1.1.67) and a fructokinase (EC 2.7.1.4), and has been identified in different bacteria, e.g., the marine Bacteroidetes Zobellia galactanivorans (Zg) which had recently gained interest to study the degradation of macroalgal polysaccharides. This protocol describes the biochemical characterization of a recombinant mannitol-2-dehydrogenase (M2DH) of Zobellia galactanivorans. The ZgM2DH enzyme catalyzes the reversible conversion of mannitol to fructose using NAD+ as a cofactor. ZgM2DH activity was assayed in both directions, i.e., fructose reduction and mannitol oxidation.
Reversible reaction:
D-mannitol + NAD+ ↔ D-fructose + NADH + H+
Materials and Reagents
- UV-Star® PS Microplate (96 Well) (Greiner Bio-One GmbH, catalog number: 655801 )
- Purified recombinant His-tagged ZgM2DH
Note: This protein was produced in Escherichia coli BL21 (DE3) containing the recombinant pFO4_ZgM2DH vector, as described by Groisillier et al. (2010). This recombinant protein was purified by affinity chromatography using a HisPrep FF 16/10 column (GE Healthcare Dharmacon) onto an Äkta avant system (GE Healthcare Dharmacon). The complete purification protocol is described in details in Groisillier et al. (2015). - Trizma® base (Sigma-Aldrich, catalog number: T1503 )
- MilliQ water
- 4-morpholineethane-sulfonic acid (MES) (Sigma-Aldrich, catalog number: M2933 )
- Bis-Tris (Sigma-Aldrich, catalog number: B9754 )
- Examples of chemicals to be tested to assess substrate and co-factor specificity:
- D-(−)-fructose (Sigma-Aldrich, catalog number: F0127 )
- D-(+)-glucose (Sigma-Aldrich, catalog number: G8270 )
- D-(+)-mannose (Sigma-Aldrich, catalog number: M4625 )
- D-(+)-galactose (Sigma-Aldrich, catalog number: G0750 )
- D-(+)-xylose (Sigma-Aldrich, catalog number: X1500 )
- D-mannitol (Sigma-Aldrich, catalog number: M9647 )
- D-sorbitol (Sigma-Aldrich, catalog number: S1876 )
- D-(+)-arabitol (Sigma-Aldrich, catalog number: A3381 )
- β-Nicotinamide adenine dinucleotide hydrate (β-NAD) (Sigma-Aldrich, catalog number: N1636 )
- β-Nicotinamide adenine dinucleotide phosphate hydrate (β-NADP) (Sigma-Aldrich, catalog number: N5755 )
- β-Nicotinamide adenine dinucleotide, reduced disodium salt hydrate (β-NADH) (Sigma-Aldrich, catalog number: N8129 )
- β-Nicotinamide adenine dinucleotide phosphate, reduced tetra (cyclohexylammonium) salt (β-NADPH) (Sigma-Aldrich, catalog number: N5130 )
- D-(−)-fructose (Sigma-Aldrich, catalog number: F0127 )
- 1 M Tris-HCl (pH 7.5) (see Recipes)
- 10 mM NADH (see Recipes)
- 10 mM NAD+ (see Recipes)
Equipment
- Safire2 UV spectrophotometer microplate reader (Tecan Trading AG)
- NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific)
Software
- Hyper 32 (hyper32.software.informer.com)
- Microsoft Excel
Procedure
- The standard fructose reduction reaction mixture contains 100 mM Tris-HCl (pH 6.5), 1 mM fructose, 0.2 mM NADH and 1 to 10 µg of purified recombinant enzyme, in a final volume of volume 100 µl. The standard mannitol oxidation reaction mixture contains 100 mM Tris-HCl (pH 8.5), 1 mM mannitol, 0.5 mM NAD+ and 1 to 10 µg of purified enzyme in 100 µl. Blank corresponds to reaction mixture where substrate is substituted by MilliQ water (Table 1).
Table 1. Composition of blank and reaction mixture for determination of ZgM2DH activity (fructose reduction)Stock solutions µl added in blank µl added in reaction mix (test) Tris-HCl (pH 6.5) (1 M) 10 10 NADH (10 mM) 2 2 Fructose (25 mM) 0 4 ZgM2DH (1 µg/µl) 1 1 MilliQ water 87 83 - The continuous assay reaction is started by adding the substrate and the activity is monitored by following changes in absorbance at 340 nm, which corresponds to the decrease or production of NADH, in a Safire2 UV spectrophotometer microplate reader. The reaction is performed at 25 °C for up to 20 min. Only the early linear part of the curves is used to calculate activity (Figure 2).
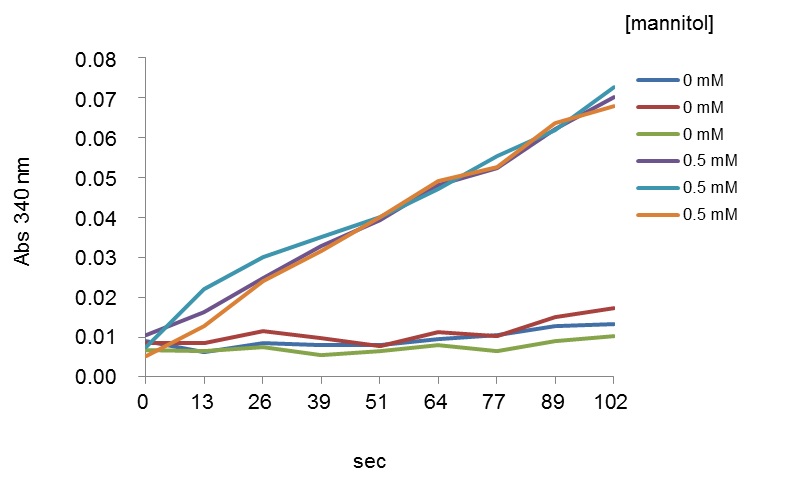
Figure 2. Early linear parts of curves representative absorbance (340 nm) monitored as a function of time (sec) in presence of mannitol. The curves represent two series of triplicates at 0 and 0.5 mM of mannitol, respectively. - Fructose reduction and mannitol oxidation activities, based on decrease or production of NADH respectively, are calculated using the formula:
[(ΔA340nm test-ΔA340nm blank)/(t*6220*0.3*0.0001)]
Where
ΔA340nm= variation of absorbance during the duration of incubation
t= duration of incubation (min)
6220= extinction coefficient for NADH (L mol-1 cm-1)
0.3= optical path (cm)
0.0001= assay volume (L)
One unit (U) of both activities (fructose reduction or mannitol oxidation) corresponds to 1 µmol of NADH reduced or oxidized per min. - To calculate specific activities, divide the value obtained in the equation above by the quantity of ZgM2DH proteins added in the reaction mixture. Perform three replicates for each assay, and determine the average ± S. E. (cf. Notes) for these three replicates. This applies also to the experiments described below. As an example, typical value for specific activity of ZgM2DH was 3.95 U/mg in presence of 10 mM fructose.
- To determine substrate specificity, test ZgM2DH activity in presence of each substrate listed in the “Materials and Reagents” section, using concentration ranging from 1 mM to at least 100 mM.
- To determine the optimal pH, replace the 100 mM Tris-HCl (pH 8.5 or 6.5) buffer used in step 1 of procedure by other buffers prepared at different pHs. As an indication, it is possible to use:
100 mM MES for pH 5.5, 6, 6.5
100 mM Bis-Tris propane for pH 6.5, 7, 7.5, 8, 8.5, 9, 9.5
100 mM Tris-HCl for pH 7, 7.5, 8, 8.5, 9 - To determine the optimal temperature, incubate the reaction mixtures used in step 1 at temperatures ranging, for instance, from 10 °C to 50 °C, with incremental of 5 or 10 °C.
- To assess the effects of various metal ions and chemicals on ZgM2DH activities, add metal ions or chemicals individually and at varying concentrations in the reaction mixture described in step 1. Examples are given in Table 2.
Table 2. Metal ions and chemicals tested for potential influence on the ZgM2DH activities. Mannitol, fructose, mannitol-1-phosphate and fructose-6-phosphate were used at high concentrations to assess potential inhibitory effects on enzymatic activity.
Compounds Final concentration in the reaction mix
(mM)Ca2+ 10 K+ 10 Li+ 10 NH4+ 10 Mg2+ 10 Mercaptoethanol 10 EDTA 10 Fructose 20 Fructose 50 Mannitol 20 Mannitol 50 Mannitol-1-phosphate 20 Fructose-6-phosphate 20 NaCl 100 NaCl 1,000 - To estimate the kinetic parameters of the enzyme for a selected substrate S, run individual enzyme reactions in presence of at least five different concentrations of this substrate and a fixed concentration of NAD(H). Determine the initial reaction rate for each reaction and plot 1/V versus 1/[S] to obtain a Lineweaver-Burk plot, from which Km and Vm for S can be calculated (Figure 3).
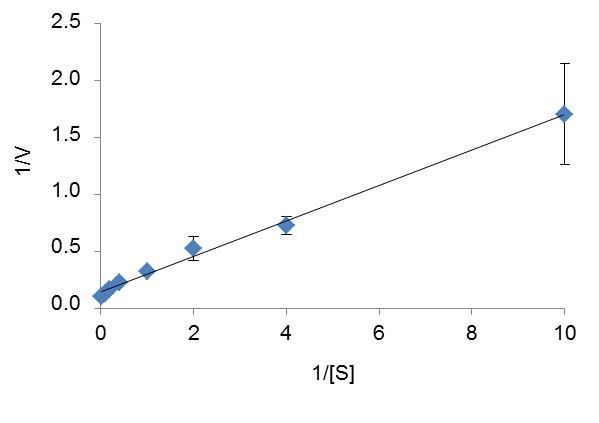
Figure 3. Lineweaver-Burk plot used to determine the Km (1.12 mM) and Vm (7.18 U/mg) of ZgM2DH for mannitol. [S] is the mannitol concentration (in mM) and V is the reaction rate (in U/mg of protein).
Notes
- Before determining Km and Vm for a given substrate S, be sure that cofactor [NAD(H)] is in excess, i.e., that V does not increase with increasing quantities of cofactor in the reaction mixture; in the same vein, before determining Km and Vm for the cofactor, be sure that S is in excess, i.e., that V does not increase with increasing quantities of S in the reaction mixture. In theory, saturating concentration is equivalent to 100 Km, but 10 Km is usually sufficient (Bisswanger, 2014). It is then necessary to adjust the ZgM2DH concentration and the incubation time to obtain a linear change of absorbance at 340 nm, i.e., a linear change of NADH.
- S. E. corresponds to standard error calculated in Excel.
Recipes
- 1 M Tris-HCl (pH 7.5)
Stored at room temperature - 10 mM NADH
Prepared fresh in water on the day of use - 10 mM NAD+
Prepared fresh in MilliQ water on the week of use and stored at -20 °C
Acknowledgments
This protocol was performed by Groisillier et al. (2015). This work was supported by the French National Research Agency via the investment expenditure program IDEALG (ANR-1 0-BTBR-02). We also acknowledge funding from the Émergence-UPMC-2011 research program.
References
- Bisswanger, H. (2014). Enzymes assays. Perspective in Sciences 1: 41-55.
- Groisillier, A., Herve, C., Jeudy, A., Rebuffet, E., Pluchon, P. F., Chevolot, Y., Flament, D., Geslin, C., Morgado, I. M., Power, D., Branno, M., Moreau, H., Michel, G., Boyen, C. and Czjzek, M. (2010). MARINE-EXPRESS: taking advantage of high throughput cloning and expression strategies for the post-genomic analysis of marine organisms. Microb Cell Fact 9: 45.
- Groisillier, A., Labourel, A., Michel, G. and Tonon, T. (2015). The mannitol utilization system of the marine bacterium Zobellia galactanivorans. Appl Environ Microbiol 81(5): 1799-1812.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Groisillier, A. and Tonon, T. (2015). Determination of Mannitol-2-dehydrogenase Activity . Bio-protocol 5(21): e1634. DOI: 10.21769/BioProtoc.1634.
Category
Microbiology > Microbial biochemistry > Carbohydrate
Biochemistry > Carbohydrate > Mannitol
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











