- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
In vitro DNA Protection Assay Using Oxidative Stress
Published: Vol 5, Iss 14, Jul 20, 2015 DOI: 10.21769/BioProtoc.1538 Views: 10240
Reviewed by: Maria SinetovaChristian RothLionel Schiavolin

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Microbial Mutagenicity Assay: Ames Test
Urvashi Vijay [...] Pradeep Bhatnagar
Mar 20, 2018 36211 Views

Colocalizing Telomeres With PML or γH2AX Foci by IF-FISH in Mouse Brain Neurons
Anna Konopka
Nov 5, 2025 1515 Views

A Quantitative DNA Fiber Assay to Monitor Replication Fork Progression, Protection, and Restart
Debanjali Bhattacharya and Ganesh Nagaraju
Feb 5, 2026 225 Views
Abstract
A wide range of stresses such as oxidative stress, acid, alkaline, UV, and metal can damage DNA. Here, we describe a protocol to measure the DNA nicking damage by Fenton reaction-mediated oxidative stress. Fenton reaction (Fe2+ + H2O2 → Fe3+ + OH- + ∙OH) produces the highly deleterious hydroxyl radicals that damage the cellular components such as DNA, lipid and proteins.
Keywords: DNA damageMaterials and Reagents
- 300 ng/µl plasmid DNA
We used 7.2 kbp pMK 3 plasmid purified from E. coli JM109 by QIA filterWe used 7.2 kbp pMK3 plasmid purified from E. coli JM109 by QIA filterTM (Plasmid Midi kit). The high quality plasmid abundant in supercoiled form is required to monitor the reduction of the supercoiled form. - Proteins to be tested on their DNA protection ability
a. BSA (Wako Pure Chemical Industries, catalog number: 01317843 )
b. Lysozyme (Wako Pure Chemical Industries, catalog number: 12202673 ) - Agarose
- Ethidium bromide (EtBr) (Invitrogen, catalog number: 15585011 )
Caution: Ethidium bromide is toxic and strong mutagen. Use appropriate gloves, safety goggles and lab coat - Ferrous ammonium sulfate (1.5 mM) (Wako Pure Chemical Industries, catalog number: 01412172 )
Note: The solution must be prepared just prior to the experiment. - 200 mM hydrogen peroxide (Wako Pure Chemical Industries, catalog number: 08104215 )
- NaCl (Wako Pure Chemical Industries, catalog number: 19101665 )
- Sodium dodecyl sulfate (SDS) (Wako Pure Chemical Industries, catalog number: 08104215)
- Tris (Nacalai tesque, catalog number: 3543421 )
- Acetic acid (Nacalai tesque, catalog number: 0021243 )
- EDTA (Nacalai tesque, catalog number: 15111 )
- CIA (chloroform:isoamyl alcohol = 24:1) (Nacalai tesque, catalog number: 0840255 )
Caution: Chloroform is suspect mutagen and harmful if inhaled. Avoid breathing vapor and prolonged contact with skin, using fume food and safety gloves. Follow the safety rule in your institute. - 300 ng/µl plasmid DNA (pMK3) (see Recipes)
- 1 µg/µl bovine serum albumin (BSA) (see Recipes)
- 1 µg/µl lysozyme (see Recipes)
- Protein/binding buffer (see Recipes)
- 1.5 mM ferrous ammonium sulfate (see Recipes)
- 200 mM hydrogen peroxide (see Recipes)
- 10% sodium dodecyl sulfate (SDS) (see Recipes)
- 1x TAE buffer (see Recipes)
- 50x TAE buffer (see Recipes)
Equipment
- Eppendorf tubes (1.5 ml)
- Eppendorf tubes (2.0 ml)
- Tips
- Thermostat bath
- Centrifuge machine
- Electrophoresis apparatus
- UV Trans illuminator and recording system (Nippon Genetics, model: e.g. FAS III system )
Software
- Image J (NIH) (http://imagej.nih.gov/ij/)
Procedure
- Pre-incubate plasmid DNA (300 ng pMK3) with or without protein sample: MrgA, MrgA*, BSA, or Lysozyme (10 and 20 µg) in 30 µl of protein-binding buffer: For 1 h at 37 °C.
- Add 8 µl of 1.5 mM fresh ferrous ammonium sulfate solution, gently mix the samples by pipetting, and then incubate for 5 min at room temperature (25 °C).
Note: Ferroxidation of ferrous iron by oxygen does not proceed significantly before the addition of hydrogen peroxide at this time scale. This 5-min incubation in our experiment is to allow MrgA to sequestrate ferrous iron in its core. - Add 2 µl of 200 mM hydrogen peroxide (final conc. 10 mM) and gently mix the samples by pipetting, and then incubate for 5 min at room temperature (25 °C).
- Add 10 µl of 10% SDS and 50 µl of CIA, and vortex. This is to denature and remove the proteins from DNA, and to recover DNA in aqueous phase in the next step.
- Recover supernatant by centrifugation at ≥13,000 x g for 5 min at 4 °C.
- Separate 20 µl of the supernatant on 1% agarose gel in TAE buffer (without EtBr) and electrophorese at 100 V for 40 min at room temperature. Following the electrophoresis, soak the gel in TAE buffer containing EtBr to visualize DNA.
- Measure the signal intensities of supercoiled DNA using NIH image J software. In detail:
- Take agarose gel images by UV-trans illuminator and CCD camera (FASIII system). Scan the photo copy by conventional scanner (if you can get high quality digital image from CCD camera, it can be directly used for the measurement. We scanned the photocopy because the resolution of the digital image is low in case of FASIII system). Measure the signal intensities of supercoiled DNA were using NIH image J software. Measure each signal intensity by subtracting the background.
- Calculate relative signal intensities using the signal intensity of the supercoiled plasmid DNA without oxidative stress.
- Calculate the mean and standard deviation of the relative intensities from multiple independent experiments. Evaluate the statistical significance by Student’s t-test.
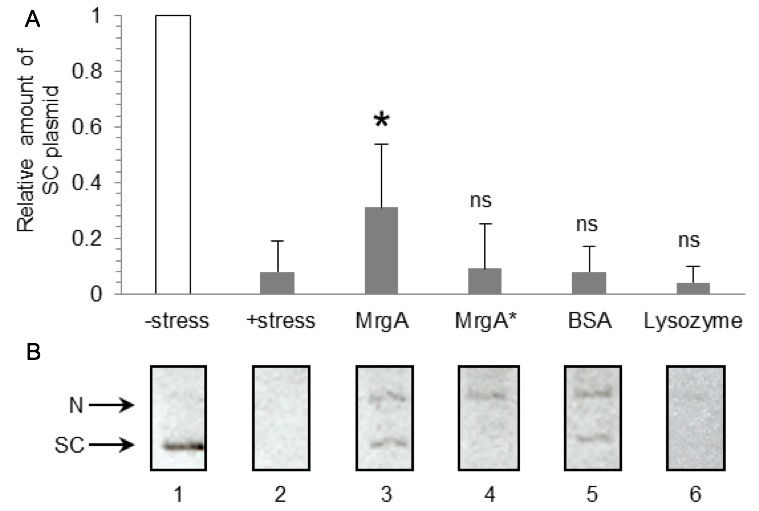
Figure 1. Representative data. A. Relative amount of supercoiled plasmid DNA. Mean and SD values of no protein (n=9), MrgA (n=4), MrgA* (n=3), BSA (n=6), and lysozyme (n=4) are shown. All proteins are 10 µg. *: Significant difference compared to oxidative stress exposure (lane 2) in t-test, p < 0.05. ns: Not significant, p >0.25. B. Representative gel image of DNA protection assay. Lanes 1: Plasmid DNA without oxidative stress. Lanes 2: Plasmid DNA treated with oxidative stress (Fe2+/H2O2). Lanes 3~6: Plasmid DNA was pre-incubated with 10 µg of MrgA (lane 3), MrgA* (lane 4), BSA (lane 5) or lysozyme (lane 6) prior to the Fe2+ addition. SC: Supercoil, N: Nicked. DNA protection by MrgA was dependent on the intact ferroxidase activity, and DNA binding by MrgA* didn’t contribute to DNA protection (Ushijima et al., 2014). - Take agarose gel images by UV-trans illuminator and CCD camera (FASIII system). Scan the photo copy by conventional scanner (if you can get high quality digital image from CCD camera, it can be directly used for the measurement. We scanned the photocopy because the resolution of the digital image is low in case of FASIII system). Measure the signal intensities of supercoiled DNA were using NIH image J software. Measure each signal intensity by subtracting the background.
Notes
- We confirmed that at least Tris or HEPES based buffer can be used for the DNA nicking damage by Fenton reaction-mediated oxidative stress.
- 1.5 mM ferrous ammonium sulfate should be freshly prepared and the dissolved solution should not exceed pH 7.0 (pH of MilliQ water is 5.50-6.86). The solution with pH values over 7.0 oxidizes ferrous iron to ferric iron immediately (Morgan and Lahav, 2007).
- Note that signal intensities can vary depending on experiments: some of the signal intensities in Figure 1B representative image are different from the mean intensities in Figure A. At least three independent experiments and the statistical evaluation are recommended. To compare distinct gel images, put reference samples that can be used to normalize the signal intensities, e.g. marker DNAs with known quantities. We employed one-side Student’s t-test.
- EDTA can be used to stop the Fenton reaction. However, when we tried to add EDTA before purification, it did not increase the DNA signal intensity, but rather generated smear signal.
Recipes
- 300 ng/µl plasmid DNA (pMK3)
Extract pMK3 purified from E. coli JM109 by Plasmid Midi Kit
Adjust concentration with MilliQ water - 1 µg/µl bovine serum albumin (BSA)
10 mg BSA
Adjust total volume to 10 ml with protein/binding buffer - 1 µg/µl lysozyme
10 mg lysozyme
Adjust total volume to 10 ml with protein/binding buffer - Protein/binding buffer
20 mM Tris-HCl (pH 8.0)
200 mM NaCl - 1.5 mM ferrous ammonium sulfate
1.17mg Ferrous ammonium sulfate
2 ml MilliQ water - 200 mM hydrogen peroxide
10.2 µl Hydrogen peroxide (30% w/v)
439.8 µl MilliQ water - 10% sodium dodecyl sulfate (SDS)
10 g sodium dodecyl sulfate
Adjust total volume to 100 ml with MilliQ water - 1x TAE buffer
20 ml 50x TAE
980 ml MilliQ water - 50x TAE buffer
242 g Tris
57.1 ml acetic acid
100 ml 0.5 M EDTA
Adjust total volume to 1 L with MilliQ water
Acknowledgments
This protocol was modified from in vitro DNA damage assay described previously (Martinez and Kolter, 1997).
References
- Martinez, A. and Kolter, R. (1997). Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J Bacteriol 179(16): 5188-5194.
- Morgan, B. and Lahav, O. (2007). The effect of pH on the kinetics of spontaneous Fe(II) oxidation by O2 in aqueous solution--basic principles and a simple heuristic description. Chemosphere 68(11): 2080-2084.
- Ushijima, Y., Ohniwa, R. L., Maruyama, A., Saito, S., Tanaka, Y. and Morikawa, K. (2014). Nucleoid compaction by MrgA(Asp56Ala/Glu60Ala) does not contribute to staphylococcal cell survival against oxidative stress and phagocytic killing by macrophages. FEMS Microbiol Lett 360(2): 144-151.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Ushijima, Y., Ohniwa, R. L. and Morikawa, K. (2015). In vitro DNA Protection Assay Using Oxidative Stress. Bio-protocol 5(14): e1538. DOI: 10.21769/BioProtoc.1538.
Category
Molecular Biology > DNA > DNA damage and repair
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









