- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Enzymatic Activity Assays for Base Excision Repair Enzymes in Cell Extracts from Vertebrate Cells
Published: Vol 5, Iss 11, Jun 5, 2015 DOI: 10.21769/BioProtoc.1493 Views: 8617
Reviewed by: Fanglian HeAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Colocalizing Telomeres With PML or γH2AX Foci by IF-FISH in Mouse Brain Neurons
Anna Konopka
Nov 5, 2025 1515 Views

Detecting the Activation of Endogenous Small GTPases via Fluorescent Signals Utilizing a Split mNeonGreen: Small GTPase ActIvitY ANalyzing (SAIYAN) System
Miharu Maeda and Kota Saito
Jan 5, 2026 496 Views

A Quantitative DNA Fiber Assay to Monitor Replication Fork Progression, Protection, and Restart
Debanjali Bhattacharya and Ganesh Nagaraju
Feb 5, 2026 225 Views
Abstract
We previously reported enzymatic activity assays for the base excision repair (BER) enzymes DNA polymerase β (pol β), aprataxin (APTX), and flap endonuclease 1 (FEN1) in cell extracts from Saccharomyces cerevisiae (Çağlayan and Wilson, 2014). Here, we describe a method to prepare cell extracts from vertebrate cells to investigate these enzymatic activities for the processing of the 5´-adenylated-sugar phosphate-containing BER intermediate. This new protocol complements our previous publication. The cell lines used are wild-type and APTX-deficient human lymphoblast cells from an Ataxia with Oculomotor Apraxia Type 1 (AOA1) disease patient, wild-type and APTX-null DT40 chicken B cells, and mouse embryonic fibroblast (MEF) cells. This protocol is a quick and efficient way to make vertebrate cell extracts without using commercial kits.
Materials and Reagents
- Cell lines used in this study
- The human cell lines used are the wild-type C2ABR and AOA1 L938 (Harris et al., 2009). The AOA1 cell line was derived from the peripheral blood of a Japanese AOA1 patient and has a point mutation within the HIT domain of APTX involving substitution of proline for leucine at position 206.
- The DT40 cell lines are the wild-type and APTX null. The APTX null cell line has the aptx gene deletion from valine 78 onwards that inactivates APTX (Ahel et al., 2006).
- The mouse embryonic fibroblast cell lines are pol β-/- and pol β+/+ MEFs previously developed in our laboratory (Sobol et al., 1996).
- RPMI 1640 medium with glutamine (Gibco, catalog number: 11875-093 )
- DMEM high-glucose medium (HyClone, catalog number: SH30081 )
- Chicken serum (Life Technologies, catalog number 16110-082 )
- Glutamax-1 (Gibco, catalog number: 35050-061 )
- Fetal bovine serum-FBS (HyClone, catalog number: SH30910 ).
- EDTA-free protease inhibitor cocktail tablet (Roche Applied Science, catalog number: 11836170001 )
- Bio-Rad Protein Dye Reagent Concentrate (Bio-Rad Laboratories, catalog number: 500-0006 )
- EDTA (Sigma-Aldrich, catalog number: 93283 )
- Potassium chloride-KCl (Sigma-Aldrich, catalog number: P9333 )
- Sodium chloride-NaCl (Sigma-Aldrich, catalog number: S7653 )
- Glycerol (Sigma-Aldrich, catalog number: G9012 )
- Tissue-culture grade 2-mercaptoethanol (Sigma-Aldrich, catalog number: M3148 )
- Nonidet P 40-NP40 (Sigma-Aldrich, catalog number: 74385 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- Magnesium chloride-MgCl2 (Sigma-Aldrich, catalog number: M8266 )
- Formamide (Sigma-Aldrich, catalog number: F9037 )
- Bromophenol blue (Sigma-Aldrich, catalog number: B0126 )
- Xylene cyanol (Sigma-Aldrich, catalog number: X4126 )
- Dithiothreitol-DTT (Sigma-Aldrich, catalog number: D0632 )
- Sodium borohydride-NaBH4 (Sigma-Aldrich, catalog number: 247677 )
- Urea (National Diagnostic, catalog number: EC-605 )
- Trizma-base (Sigma-Aldrich, catalog number: T4661 )
- Boric Acid (Promega Corporation, catalog number: H5003 )
- AccuGel (40%) 19:1 Acrylamide to Bisacrylamide Stabilized Solution (National Diagnostic, catalog number: EC-850 )
- Ammonium persulfate (Sigma-Aldrich, catalog number: A3678-25G )
- Tetramethylethylenediamine (Sigma-Aldrich, catalog number: T9281-25ML )
- Sterile water
- Purified enzymes: Recombinant human DNA polymerase β [purified as described Çağlayan et al. (2014)], recombinant human APTX (Fitzgerald catalog number: 80R-1256 ), and recombinant human FEN1 [purified as described Çağlayan et al. (2014)].
- DNA substrate: The gapped DNA substrate with a uracil base at position 17 at the 5'-end of the 3'-end FAM-labeled oligonucleotides. The sequence information for the upstream, downstream and template oligonucleotides were previously published Çağlayan et al. (2015).
- Lysis buffer (see Recipes)
- 10x reaction buffer (see Recipes)
- Gel-loading buffer (see Recipes)
- 10x TBE solution and 1x TBE solution as PAGE running buffer (see Recipes)
- 15% Denaturing Polyacrlamide Gel or PAGE solution (see Recipes)
Equipment
- Eppendorf tubes
- Screw cap conical tube (15 ml)
- Refrigerated table-top centrifuge
- Refrigerated Eppendorf centrifuge
- Table-top heat block
- Tissue culture CO2 incubators set at 34, 37, and 39.5 °C
- Cell scraper
- Polyacrylamide gel electrophoresis (PAGE) apparatus
Procedure
- Cell growth
- DT40 cells (wild-type and APTX-null) are maintained in RPMI 1640 medium supplemented with 10% FBS, 1% chicken serum and 2 mM L-glutamine. 2-mercaptoethanol (50 μM) is added fresh to the medium at the time of use and cells are grown in a 5% CO2 incubator at 39.5°C (Okamoto et al., 2014).
- The human lymphoblastoid cells (wild-type and APTX-deficient AOA1) are maintained in RPMI 1640 medium containing FBS and grown in a 5% CO2 incubator at 37°C (Harris et al., 2009).
- MEF cells (wild-type and pol β-deficient) are maintained in DMEM medium containing 10% FBS and 4 mM glutamax and grown in a 10% CO2 incubator at 34°C (Sobol et al., 1996).
- 1-2 x 150 mm dishes of MEF cells are washed twice in 10 ml cold PBS, then harvested by scraping. Cells are collected in 15 ml PBS in a 15 ml tube, then centrifuged at 1600 rpm at 4 °C for 5 min.
- The required volume of suspension cells (DT40 and human lymphoblasts) is centrifuged at 1600 rpm at 4 °C for 5 min, and the cell pellet are transferred to a 15 ml tube and washed twice with 10 ml PBS.
- Cell pellets are transferred to Eppendorf tubes in 1 ml PBS and briefly spun down. Supernatants are discarded and pellets are frozen immediately in dry ice and stored at -80 °C until use.
- Pellets of 20-50 million cells are used for cell extract preparation.
- DT40 cells (wild-type and APTX-null) are maintained in RPMI 1640 medium supplemented with 10% FBS, 1% chicken serum and 2 mM L-glutamine. 2-mercaptoethanol (50 μM) is added fresh to the medium at the time of use and cells are grown in a 5% CO2 incubator at 39.5°C (Okamoto et al., 2014).
- Preparation of cell extracts
- The cell extracts from vertebrate cells are prepared as reported (Biade et al., 1998) and summarized below.
- Resuspend the cell pellet in 400 μl of ice-cold lysis buffer.
- Rotate the resuspended cell pellets for 1 h at 4 °C.
- Centrifuge the mixture at 14,000 rpm at 4 °C for 10 min to remove cell debris.
- Carefully transfer the supernatant fraction to a fresh Eppendorf tube. Be careful not to touch the pellet.
- Determine the protein concentration of the extract using Bradford assay dye reagent with BSA as standard (Bradford, 1976).
- The cell extracts from vertebrate cells are prepared as reported (Biade et al., 1998) and summarized below.
- Enzymatic activity assays in cell extracts
- Prepare 10 μl of reaction mixture (final volume) including 1x reaction buffer and 100 nM DNA substrate. The DNA substrate used includes an adenylated uracil base at the 5′ end of the 3′-FAM–labeled oligonucleotide (Çağlayan et al., 2014). For the reference reactions including purified proteins, pol β, APTX, and FEN1, the reaction mixture included a gapped DNA substrate that was pre-incubated with UDG as described (Çağlayan et al., 2014). For the reactions including cell extracts, the DNA substrate was included in the reaction mixture without UDG pretreatment.
- Start the reaction by adding cell extract, prepared as above, to the reaction mixture. For the reference reactions, start each reaction by adding the purified protein to the reaction mixture in final concentrations as follows: pol β (500 nM), APTX (100 nM), and FEN1 (100 nM) (Çağlayan et al., 2014 and Çağlayan et al., 2015).
- Incubate the reaction mixture at 37 °C for 15 min.
- Stabilize the reaction products by addition of 1 M freshly prepared and ice-cold NaBH4 to a final concentration of 100 mM.
- Incubate the reaction samples on ice for 30 min.
- Mix the reaction products with 10 μl of gel-loading dye.
- The reaction products in cell extracts from vertebrate cells are separated on a 15% polyacrylamide gel. The gel is scanned, and the data analyzed as reported (Çağlayan et al., 2014 and Çağlayan et al., 2015).
- Prepare 10 μl of reaction mixture (final volume) including 1x reaction buffer and 100 nM DNA substrate. The DNA substrate used includes an adenylated uracil base at the 5′ end of the 3′-FAM–labeled oligonucleotide (Çağlayan et al., 2014). For the reference reactions including purified proteins, pol β, APTX, and FEN1, the reaction mixture included a gapped DNA substrate that was pre-incubated with UDG as described (Çağlayan et al., 2014). For the reactions including cell extracts, the DNA substrate was included in the reaction mixture without UDG pretreatment.
Representative data
A sample gel image of products for the pol β dRP lyase, FEN1 excision and APTX DNA deadenylation enzymatic activities in the extracts from DT40 cells is presented below (Figure 1). Migration positions of DNA substrate (line 1) and the reaction products after pol β lyase removal of the 5’-AMP-dRP group (indicated as 17-mer and shown at line 2 in reference reaction and lines 5-6 in cell extracts), APTX 5’-AMP removal (indicated as 17dRP, and shown at line 3 in reference reaction and lines 6 in cell extract), and FEN1 excision products (indicated as 15-mer and 16-mer and shown at line 4 in reference reaction and lines 5-6 in cell extracts) are presented. The original results showing enzymatic activities in the cell extracts from the vertebrate cell lines listed below were published in Çağlayan et al. (2015).
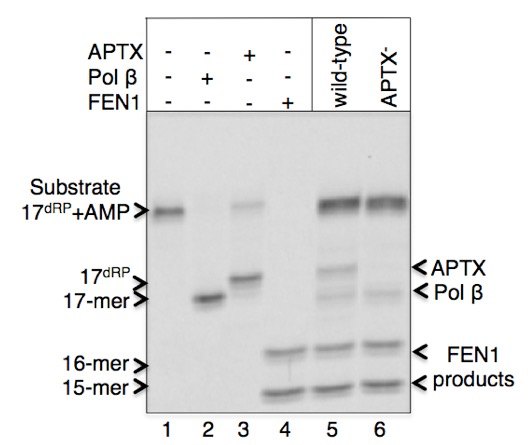
Figure 1.Pol β, APTX, and FEN1 activities in cell extracts from DT40 wild-type and APTX-null cell lines
Notes
After addition of lysis buffer to the cell pellet, the Eppendorf tube including resuspended cells should be gently mixed by tapping. Enzymatic activity in vertebrate cell extracts may be lost if samples are mixed by vortex instead of rotating at 4 °C. Typically, the enzymatic activities are determined under steady-state conditions with the substrate in excess over enzyme and under conditions of a linear time-course. In addition, the enzymatic activity should be under conditions of linearity between activity and amount of extract added to the reaction mixture.
Recipes
- Lysis buffer
10 mM Tris–HCl (pH 7.8 in the final buffer solution)
200 mM KCl
1 mM EDTA
20% glycerol
0.1% NP-40
1 mM DTT (add fresh)
Protease inhibitor tablet
- 10x reaction buffer
500 mM HEPES (pH 7.5 in the final buffer solution)
100 mM MgCl2
200 mM KCl
5 mM EDTA
20 mM DTT
- Gel-loading buffer
95% formamide
20 mM EDTA
0.02% bromophenol blue
0.02% xylene cyanol
- 10x TBE solution
108 g Tris-base
55 g Boric Acid
40 ml 0.5 M EDTA
H20 up to 1 L
- Denaturing PAGE solution (15%)
40 g Urea
30 ml 19:1 AccuGel (40%)
8 ml 10% TBE solution
10 ml H20
250 μl 10% APS
35 μl TEMED
Acknowledgments
This work was supported by the Intramural Research Program of the US National Institutes of Health, National Institute of Environmental Health Sciences (project numbers Z01 ES050158 and ES050159).
References
- Ahel, I., Rass, U., El-Khamisy, S. F., Katyal, S., Clements, P. M., McKinnon, P. J., Caldecott, K. W. and West, S. C. (2006). The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature 443(7112): 713-716.
- Biade, S., Sobol, R. W., Wilson, S. H. and Matsumoto, Y. (1998). Impairment of proliferating cell nuclear antigen-dependent apurinic/apyrimidinic site repair on linear DNA. J Biol Chem 273(2): 898-902.
- Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248-254.
- Çağlayan, M. and Wilson, S. H. (2014). Enzymatic activity assays in yeast cell extracts. Bio-protocol 4(23): e1312.
- Caglayan, M., Batra, V. K., Sassa, A., Prasad, R. and Wilson, S. H. (2014). Role of polymerase beta in complementing aprataxin deficiency during abasic-site base excision repair. Nat Struct Mol Biol 21(5): 497-499.
- Caglayan, M., Horton, J. K., Prasad, R. and Wilson, S. H. (2015). Complementation of aprataxin deficiency by base excision repair enzymes. Nucleic Acids Res 43(4): 2271-2281.
- Harris, J. L., Jakob, B., Taucher-Scholz, G., Dianov, G. L., Becherel, O. J. and Lavin, M. F. (2009). Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum Mol Genet 18(21): 4102-4117.
- Okamoto, S., Narita, T., Sasanuma, H., Takeda, S., Masunaga, S., Bessho, T. and Tano, K. (2014). Impact of DNA repair pathways on the cytotoxicity of piperlongumine in chicken DT40 cell-lines. Genes Cancer 5(7-8): 285-292.
- Sobol, R. W., Horton, J. K., Kuhn, R., Gu, H., Singhal, R. K., Prasad, R., Rajewsky, K. and Wilson, S. H. (1996). Requirement of mammalian DNA polymerase-beta in base-excision repair. Nature 379(6561): 183-186.
Article Information
Copyright
© 2015 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Çağlayan, M., Horton, J. K. and Wilson, S. H. (2015). Enzymatic Activity Assays for Base Excision Repair Enzymes in Cell Extracts from Vertebrate Cells. Bio-protocol 5(11): e1493. DOI: 10.21769/BioProtoc.1493.
Category
Molecular Biology > DNA > DNA damage and repair
Biochemistry > Protein > Activity
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










